PATH-SAFE
PATH-SAFE is a 4-year, UK wide and cross government programme, led by the FSA and utilising £24m funding from the HMT Shared Outcomes Fund (SOF) and match funding from a range of government and academic delivery partners.
PATH-SAFE was a 4-year, cross government and UK wide programme, led by the Food Standards Agency (FSA) that utilised £24m of funding from the HMT Shared Outcomes Fund (SOF) and match funding from a range of government and academic delivery partners. The programme piloted the development of a national surveillance network, using the latest DNA-sequencing technology and environmental sampling, to improve the detection and tracking of foodborne human pathogens and associated AMR through the whole agri-food system from farm-to-fork.
PATH-SAFE ended in March 2025, however this page will continue to be updated with links to programme outputs, and will transition to include information about follow on biosurveillance work in due course.
In October 2024, a commentary summarising the program's achievements, and consideration on how they could be taken forward beyond the lifetime of the program, was published in Future Microbiology (October 2024): Coordinated surveillance of foodborne pathogens and antimicrobial resistance: insights from the PATH-SAFE pilot.
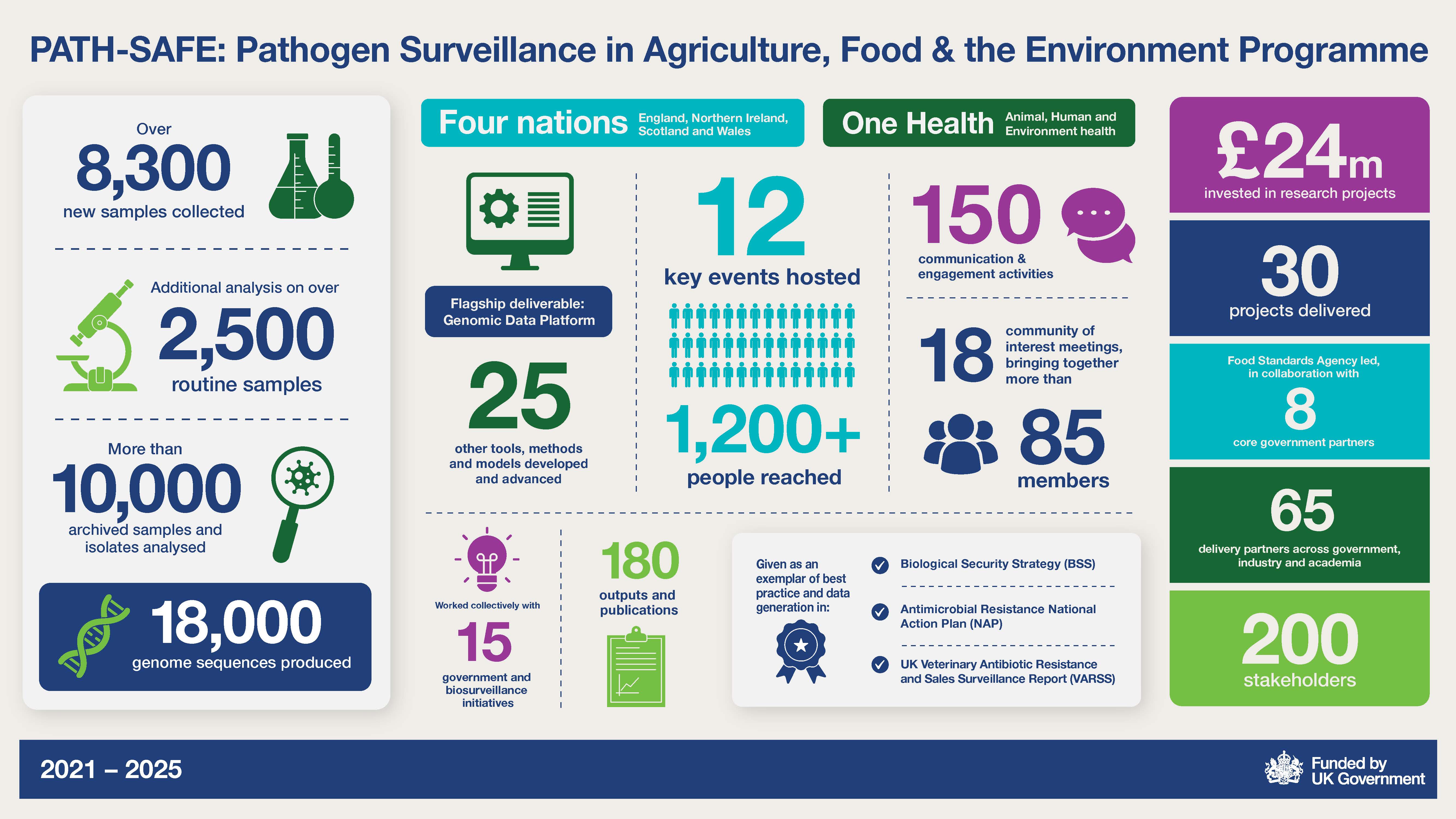
£24 million invested in research projects. 30 projects delivered. Food Standards Agency led, in collaboration with 8 core government partners. 65 delivery partners across government, industry and academia. 200 stakeholders. 2021 to 2025: Over 8300 new samples collected. Additional analysis on over 2500 routine samples. More then 10000 archived samples and isolates analysed, 18000 genome sequences produced. Four nations - England, Northern Ireland, Scotland and Wales. One Health - Animal, Human and Environment health. Flagship deliverable: genomic data platform 25 other tools, methods and models developed and advanced. 12 key events hosted. 1200 people reached. 150 communication and engagement activities. 18 community of interest meetings, bringing together more than 85 members. Worked collectively with 15 government and biosurveillance initiatives. 180 outputs and publications. Given as an exemplar of best practice and data generation in Biological Security Strategy (BSS), Antimicrobial Resistance National Action Plan (NAP), UK Veterinary Antibiotic Resistance and Sales Surveillance Report (VARSS)
Programme background
Foodborne disease (FBD) is a major public health risk with 2.4 million individual illnesses and more than 16,000 hospitalisations per year. Most human disease is caused by a handful of bacteria which enter the food chain from farmed animals or the environment.
In addition to FBD, the agri-food supply chain also poses a risk for the transmission of antimicrobial resistance (AMR) through food, animals, humans, and water. Whilst the UK has made progress in reducing its use of antibiotics in humans and animals in the last five years, drug-resistant bloodstream infections in humans have increased by 32% from 2015 to 2019. The rise and spread of AMR is creating a new generation of ‘superbugs’ that cannot be treated with existing medicines.
For these reasons, government departments already undertake surveillance activities by analysing samples from food, livestock, and humans. Advances in technology and data management offer the opportunity to create a step change in surveillance, to protect public health. The PATH-SAFE programme established a new data platform that allows for the analysis, storage and sharing of pathogen sequence and source data, collected from multiple locations across the UK by government departments and public organisations. This single system will enable rapid identification and tracking of FBD and AMR. This will improve public health and minimise the economic and environmental impact of outbreaks.
Aims of the programme
- To pilot a better national surveillance system for the monitoring and tracking of FBD and AMR in the environment and agri-food system
- To bring together and build on existing initiatives across the UK and to understand what the end-user needs to improve how they work in this space
- To provide better data to identify the prevalence, source and pathways of FBD and AMR, helping to prevent spread by enhanced targeting of interventions
Benefits of the programme
- Generation of information and knowledge that support enhanced identification of FBD and AMR sources
- Providing improved user access to relevant data so they can make more informed, evidence-based decisions
- Data that underpins the prevention and prediction of FBD and AMR spread by supporting improved, cost-effective targeting of interventions, providing economic savings both for government and industry
- Improved surveillance system that supports better identify and quicker reaction to emergent diseases (or diseases of increasing concern).
- Potential to reduce the number and cost of FBDs and AMR, lower commercial losses, strengthen UK Science Excellence, and enhance the UK food sector’s reputation
Programme partners
The PATH-SAFE programme was led by the FSA, supported by several core partners:
- Food Standards Scotland (FSS)
- Department of Environment, Food and Rural Affairs (Defra)
- Animal and Plant Health Agency (APHA)
- Centre for Environment, Fisheries and Aquaculture Science (Cefas)
- Environment Agency (EA)
- Veterinary Medicines Directorate (VMD)
- Department of Health and Social Care (DHSC)
- UK Health Security Agency (UKHSA)
The programme was delivered through a one-health, four-nation collaborative approach, both at the delivery level and at the strategic level.
More than 65 organisations, from across government, academia and industry, have been involved in delivery and advisory roles. These have included:

Department for Environment, Food and Rural Affairs, Animal & Plant Health Agency, UK Health Security Agency, Centre for Environment, Fisheries and Aquaculture Science, Veterinary Medicines Directorate, Department of Health and Social Care, Food Standards Scotland, Scottish Environment Protection Agency, Scotland's Rural College, Public Health Scotland, Food Standards Agency, Environment Agency, Northern Ireland Environment Agency, NHS Lothian, Department of Heath (NI), Agriculture, Environment and Rural Affairs (NI), Department of Infrastructure (NI), Welsh Government, HSC Public Health Agency, Northern Ireland Water, Agri-food and biosciences institute, Public Health Wales, Welsh Water, South West Water, Ribble Rivers Trust, Biosphere North Devon, Virosafety, resistomap, Quadram Institute, Moredun Research Institute, EnteroBase, Queen's University Belfast, University of Lincoln, Cardiff University, Climb big data, The University of Edinburgh, University of Exeter, Cranfield University, Bangor University, The University of Warwick, University of Oxford, University of Birmingham, University of Worcester, UK Pet Food, Agricultural Industries Confederation, Arla, Fera, UK Centre for Ecology and Hydrology, Centre for Genomic Pathogen Surveillance, Digital Epidemiology Services Ltd, Aecom, Arup, Capita, Deloitte, National Milk Laboratories, MicrobesNG, RAND Europe, NHS Greater Glasgow and Clyde, The City of Edinburgh Council, Glasgow City Council, Met Office, Channel Costal Observatory, PubMLST, Pathogenwatch, Nijhuis
Funding source and timeline
PATH-SAFE was funded via the Treasury Shared Outcomes Fund (SOF).
Spending Round 2019 announced £200 million for the Shared Outcomes Fund to fund pilot projects to test innovative ways of working across the public sector. The first round of the SOF funded a wide range of projects to be run in 2020-21 and 2022-23.
Spending Review 2020 announced a further £200 million was to be made available for a second round of the Shared Outcomes Fund between 2021-22 and 2023-24.
In 2021 the programme successfully secured £19.2m in phase 2 of the SOF, to run a three-year programme concluding in March 2024.
At the start of 2024, PATH-SAFE successfully secured an additional £4.7m of funding to continue for an additional year (April 2024 to March 2025), with HMT SOF providing £2.2m of the continuation funding, leveraged by £2.5m of match funding from PATH-SAFE core government partner departments and academic delivery partners.
Programme themes
The programme was organised thematically for governance, oversight, reporting and coordination purposes.
Further information about each of the themes, including overviews, delivery partners, status, key successes and outputs can be found on the following pages:
- National foodborne disease genomic data platform
- Onsite Diagnostics
- Novel Foodborne Pathogen, and associated antimicrobial resistance, knowledge and surveillance approaches
- Programme Evaluation
The programme developed new systems and tools, explored novel surveillance approaches and generated knowledge, to improve the detection and tracking of foodborne human pathogens and antimicrobial resistance, through the whole agri-food system from farm-to-fork.

Alongside the technical advances the programme has driven forward, PATH-SAFE has also established and promoted a collaborative, four nation, One Health approach to surveillance, which has been referenced as an exemplar within initiatives such as the Biological Surveillance Strategy and the UK AMR National Action plan (noted within as a key achievement).
The programme has influenced, informed, and supported programmes, strategies and initiatives across government and beyond, including:
- Biothreats Radar and National Biosurveillance Network (NBN)
- UK Microbial Forensics Consortium (UKMFC)
- Genomics of Animal and Plant Disease Centre Phase 2 (GAP-DC2)
- AMR National Action plan (NAP) 2019-24 and 2024-29
- UKRI Tackling Infections Strategic Theme
- Government Office for Technology Transfer (GOTT)
- Microbiology Society – Knocking Out AMR initiative
- ONS Integrated Data Service (IDS) Programme
- Food Safety Research Network (FSRN)
Events and communications
Throughout the programme, a range of events and communication activities have been undertaken including a webinar series, a conference and publication of quarterly newsletters.
Further information on these events and activities can be found on the following pages:
Get in touch
If you would like to get in touch with the PATH-safe team, you can email them at pathsafe@food.gov.uk
Overview
The UK is a recognised global leader in genomic database systems. We are utilising this existing expertise, working with academic colleagues and major ‘big data’ stakeholders, to create a user-friendly platform for the rapid interrogation and contextualisation of genomic data.
The rapid detection, identification and tracking of pathogens has always been fundamental to public health, and due to recent advances in DNA-based methods we now have the means to do this. As noted in the UK National Data Strategy, innovation has led to an exponential growth in data - in its generation and use, and in the world’s increasing reliance upon it.
By embracing data and its benefits, the UK now faces tangible opportunities to improve our society and grow our economy. It can transform our public services and dramatically improve health outcomes nationally. Modern genome sequencing offers the potential to do just that, by identifying pathogen strains rapidly and cheaply with a high resolution, reconstruct chains of transmission and trace outbreaks to their source. However, we need the data infrastructure to properly enable this.
The PATH-SAFE genomic data platform seeks to address this need at a national level, by providing a cross-government genomics capability that can perform analysis of genomic data and associated metadata. This can facilitate rapid identification of pathogen strains of interest and support elucidation of transmission pathways.
The platform aims to realise the following benefits:
- improve the tracking of foodborne disease (FBD) pathogens and associated antimicrobial resistance (AMR) through faster identification of variants of interest, thereby increasing outbreak investigation efficiency
- through analysis, bring together currently unconnected government and public genomic pathogen data to maximise the use of available FBD biosurveillance data and thereby expedite variant identification
- create efficiencies in cross-government working in FBD biosurveillance through data sharing and collaboration via the platform
- reduce public health risks associated with foodborne disease outbreaks by tracing sources of infection faster, potentially facilitating faster removal of those sources
- reduce economic losses associated with FBD by reducing the number of missed, slow or inaccurate assessments of outbreak cause, thereby reducing the costs and resources associated for people affected by FBD (such as sick days) and by businesses (such as additional administrative costs of dealing with an outbreak)
Delivery partners
Our delivery partners for this platform are:
- Digital Epidemiology Services
- University of Oxford
- University of Warwick
- University of Birmingham
Key successes
Key success of this work include:
- following a 15-month development period, the first iteration of the genomic data platform was made live in March 2024, ready for submission and analysis of Salmonella genomes by participating government partner agencies
- the platform and its functionality has built upon existing and proven surveillance tools and infrastructure, with CLIMB-BIG-DATA providing the cloud infrastructure used by the platform, Pathogenwatch tools supplying the platform’s analytics capability and a call-out feature to Enterobase supporting the platform’s context genome search functions
- a discovery phase was completed prior to development being initiated, which sought to clarify existing surveillance infrastructure and processes to inform platform design
- extensive end user engagement undertaken during platform development, ahead of release of the first iteration of the platform (another opportunity will be provided ahead of the second iteration)
- set-up of four advisory groups (Technical, AMR, Data Standards and International), which provided recommendations on the design and capabilities of the platform and on the application, technical requirements and best practices for whole genome sequencing (WGS) in Salmonella
- FBD surveillance in general - a recommendations paper has been collated and is due for publication in 2025
Outputs
All projects were presented at the PATH-SAFE Biosurveillance Conference on 28 and 29 February 2024 in London. Recordings, slides and posters are available on the conference webpage.
Outputs will continue to be added here throughout 2025 and 2026. If the information you're looking for isn't currently available, please check back again soon.
Overview
The advancement of rapid and accurate onsite diagnostic methods provides a real opportunity in relation to detection and control of foodborne pathogens.
The first phase of this work undertaken in 2022/2023 and 2023/2024 focused on the identification of promising technologies that could be piloted for onsite testing of foodborne pathogens, including piloting selected technologies.
A comprehensive literature review was undertaken to identify technologies under development for on-site testing. This included technologies proven or presumed to be applicable in detecting targets relevant to food safety, as well as those developed for other sectors. These technologies were assessed through a customised technology readiness level (TRL) framework with each technology assigned a TRL in combination with each in-scope pathogen or matrix.
To understand the testing requirements in real world scenarios, both "strategic" and "operational" stakeholders were mapped and engaged in focus groups and interviews. This provided a broad perspective on on-site testing and supported identification of sectors or processes that are well-suited or not suited for portable detection technologies.
Results from the literature review and end-user study led to the shortlisting of technologies based on relative maturity and suitability for onsite use, mapped against potential applications within the food sector for deployment of the technologies. This decision matrix allowed an evaluation of each of the shortlisted technologies in the context of specific end-user needs, and a final selection was made based on the outcome.
Two technologies were selected, validated in the laboratory and various performance criteria such as specificity, sensitivity, repeatability, and reproducibility were evaluated. The finalised methods were tested in the hands of end-users, with end-user training initially provided in a controlled environment and subsequently the end users conducted the tests themselves onsite and provided feedback.
A separate, but related piece of work was undertaken by UKHSA and 20/30 Labs to explore the possibility of repurposing rapid, Covid-19 in-field wastewater diagnostic technology for the detection of foodborne pathogens. This work demonstrated that rapid detection of seven foodborne pathogens in wastewater is feasible using rapid Loop-mediated isothermal amplification (LAMP) based technologies.
This work has demonstrated the potential utility of using onsite diagnostics as inspection tools in the foodborne disease (FBD) arena, whilst also identifying that there are no 'ready-made' solutions on the market that meet the specific needs of end-users but found that there are a wide variety of emerging technologies. It has therefore become apparent that guidance for the ‘real world’ deployment of onsite diagnostics is essential. The focus of the 24/25 continuation work is to develop a set of guidelines, in particular for the use of onsite diagnostics for official controls in the food sector. End user insights and perspectives will be sought, and a case study will be developed to illustrate application of the guidelines. Needs from the policy, legislative and accreditation body perspectives will also be considered (Fera).
Delivery partners
Delivery partners for this work:
- Fera Science
- University of Lincoln
Key successes
Key successes for this work include:
- horizon scanning identified more than 65 potential onsite diagnostic technologies, which were subsequently assessed using a Technology Readiness Level (TRL) framework developed as part of PATH-SAFE
- two onsite diagnostic technologies successfully piloted by end users in onsite locations and key scenarios
- significant end-user engagement through the completion of an end-user study to understand community needs and to help in identifying sectors or processes that are well-studied, or not, for portable detection technologies
Outputs
All projects were presented at the PATH-SAFE Biosurveillance Conference on 28 and 29 February 2024 in London. Recordings, slides and posters are available on the conference webpage.
Overview
The work within this theme has focused on three key areas:
- development of novel surveillance methodologies, approaches and tools
- generation of data on the genomic diversity of a range of foodborne pathogens, and associated antimicrobial resistance (AMR), across the four nations of the UK to establish baselines and address knowledge gaps
- exploration of whole genome sequence data to investigate foodborne pathogen and antimicrobial resistance transmission routes
Collaborating with a range of delivery partners and drawing on their strength and expertise has enabled the delivery of a large portfolio of projects, which have incorporated perspectives and input from across the agri-food system.
Food Standards Scotland (FSS) have sequenced thousands of Escherichia coli (E.coli) isolates from a diverse range of sources across Scotland. This will help elucidate transmission dynamics through the agri-food chain and develop machine learning source attribution models for predicting the source or host of origin for E.coli isolates. The team are now sequencing further agri-food related isolates to verify the application of the E. coli source attribution models, looking to demonstrate proof of concept for use of the model in the investigation of foodborne illness and risk management, and extend the approach to develop a similar model for Salmonella Typhimurium (STm).
The Animal and Plant Health Agency (APHA) and Veterinary Medicines Directorate (VMD), have generated new data and knowledge on AMR presence, diversity, and transmission by applying genomics to existing surveillance sampling and initiating novel sampling in understudied compartments of the agri-food chain, particularly ruminants (via bulk milk, and cattle and sheep at slaughter) and animal feed. Whole genome sequence data produced by these projects is now undergoing enhanced genomic analysis to enhance understanding of AMR dynamics, transmission pathways and mobile genetic elements dissemination. Further novel sampling is being conducted to determine the prevalence of AMR bacteria in imported raw pet food, and to explore the application of novel abattoir environment sampling approaches as a potential alternative surveillance method that could act as a proxy for existing labour-intensive livestock sampling methods.
The Environment Agency (EA) have developed a pilot environmental surveillance system for AMR in the environment, including river water and bathing waters, wild flora and fauna, shellfish and bioaerosols. This work is continuing as part of the Environment Agency research portfolio.
The Centre for Environment, Fisheries and Aquaculture Science (Cefas) undertook a river catchment pilot to compare wastewater, river water and shellfish for monitoring of a range of foodborne pathogens, and have worked with UKHSA and the Public Analyst laboratories in Scotland to demonstrate the utility of wastewater for monitoring norovirus and salmonella on a national scale. The team is now continuing the application and advancements of methodologies developed in PATH-SAFE to demonstrate the utility of bivalve molluscan shellfish (BMS), a species routinely sampled through pre-existing surveillance frameworks, as a sentinel in a ‘sample once test many’ approach.
Bangor University are examining the flow, dynamics and risk posed by AMR and foodborne pathogens (FBP) in wastewater discharges, particularly those originating from healthcare settings. Further work is ongoing to investigate seasonal patterns and potential wastewater treatment opportunities and barriers.
Food Standards Agency (FSA), UK Health Security Agency (UKHSA), NHS Lothian and Cefas are undertaking a project to generate foodborne disease (FBD) prevalence and WGS data from business-as-usual UKHSA polio wastewater monitoring, utilising methodologies developed by Cefas within their PATH-SAFE project. This data will subsequently be compared to data from clinical samples collected through the third study of Infectious Intestinal Disease in the UK, presenting an exciting opportunity to align two large surveillance efforts to generate insights into the effectiveness of wastewater monitoring for FBP.
Queen’s University Belfast (QUB) have been investigating the application of air and near-source wastewater monitoring for AMR and FBP in a care home setting in Northern Ireland. This work is continuing to explore seasonal trends, increase understanding of AMR and foodborne pathogens prevalence and transmission, and further evidence the utility of the novel surveillance methods.
University of Oxford have sequenced thousands of Campylobacter isolates from a diverse range of collaborators, to help elucidate source attribution and transmission dynamics through the agri-food chain and the rise in AMR. Sequencing and analysis of further human and agri-food Campylobacter isolates is ongoing to determine levels of AMR in Campylobacter isolates currently circulating in the UK, and enhance understanding of AMR gene transmission.
Agri-food and Biosciences Institute (AFBI) in Northern Ireland selected and sequenced archived isolates of Salmonella and Listeria monocytogenes from food and animal sources, to provide important historical context against which future outbreak sequences can be compared.
Delivery partners
In addition to the lead delivery partners highlighted in the 'Overview' section, a number of other collaborators have supported the delivery of these projects, including:
- Scottish Environment Protection Agency
- Public Health Scotland
- Scotland's Rural College
- Moredun Research Institute
- The University of Edinburgh
- Northern Ireland Environment Agency
- Department of Health (NI)
- Agriculture, Environment and Rural Affairs (NI)
- Department of Infrastructure (NI)
- HSC Public Health Agency
- Northern Ireland Water
- Welsh Government
- Public Health Wales
- Welsh Water
- Cardiff University
- South West Water
- Ribble Rivers Trust
- Biosphere North Devon
- Virosafety
- Resistomap
- Quadram Institute
- University of Exeter
- Cranfield University
- UK Pet Food
- Agricultural Industries Confederation
- National Milk Laboratories
- Arla
- UK Centre for Ecology and Hydrology
- Aecom
- Arup
- Capita
- Deloitte
- MicrobesNG
Key successes
Across the theme:
- more than 8,300 samples have been collected, across a range of sample sources and four nations
- additional analysis has been undertaken on over 2,500 samples collected as part of existing surveillance activities
- exploration of more than 10,000 archived samples and pathogen isolates
- more than 18,000 pathogen isolates have been sequenced
- increased understanding of methods suitable for environmental monitoring across a range of different media, and development of a pilot environmental surveillance system
Novel surveillance methods, approaches and tools developed and tested using real world samples and data for the first time, including:
- protocols for isolation and testing of foodborne pathogens from river water, wastewater and shellfish
- wastewater-based AMR and FBD surveillance sample collection schedules, sample analysis and data analysis pipelines
- machine learning models to determine source/host attribution of pathogens
Contamination and transmission models:
- demonstrable benefit of wastewater-based surveillance, inspiring nursing homes in Northern Ireland to consider wastewater systems design in future builds and support wastewater-based surveillance
- large sample volume, broad scope and high throughput nature of PATH-SAFE projects have accelerated development and refinement of existing and established methods, including long read sequencing methodologies used by APHA for routine AMR surveillance
- utility of routine whole genome sequencing demonstrated through patterns, trends and insight gained across the programme, including the identification of resistant bacterial isolates of concern within the isolates sequenced for PATH-SAFE
- strengthened cross-government and academia collaborative approach to identifying and pursuing surveillance opportunities, methods development and results analysis
- industry actively involved in the design and conduct of government surveillance activities and promoting their benefits, including milk producers and an animal feed mill, in collaboration with National Milk Record and The Agricultural Industries Confederation respectively
Outputs
All projects were presented at the PATH-SAFE Biosurveillance Conference on 28 and 29 February 2024 in London. Recordings, slides and posters are available on the conference webpage.
Summary articles
Monitoring antimicrobial resistance in the environment. Published 24 October 2024 on gov.uk, a piece within the Environment Agency's Creating a better place blog.
How the PATH-SAFE programme has driven forward our understanding of AMR in UK animals: Published 18 November 2024 in The Microbiologist, Applied Microbiology International's digital magazine. This piece highlights how evidence generated within PATH-SAFE has progressed the understanding of AMR prevalence and transmission, and illustrates the significant benefits that well-funded, coordinated, cross-sectoral initiatives can deliver.
Environment Agency publications
Scoping review into environmental selection for antifungal resistance and testing methodology. Published on 22 July 2022 on gov.uk, this report reviews the current understanding of the mechanism for selection for antifungal resistance in fungal species following exposure to antifungals.
Antimicrobial resistance surveillance pilot site selection and database extension. Published on 22 July 2022 on gov.uk, this project developed selection criteria to identify suitable river catchments for piloting a surveillance programme for environmental antimicrobial resistance.
Sampling strategy and assessment options for environmental antimicrobial resistance in airborne microorganisms. Published on 22 July 2022 on gov.uk, this report reviews the available sampling options for antimicrobial resistant microorganisms, including their antimicrobial resistance genes, from the atmosphere.
Antifungal medicines in the terrestrial environment: Levels in biosolids from England and Wales. Published on 02 February 2023 in Science of The Total Environment, this article reports on biosolids from ten sites in England and one in Wales that were sampled and analysed for clinical antifungals.
Environmental surveillance of AMR, perspective from a national environmental regulator in 2023. Published 30 March 2023 in Eurosurveillance, this is a perspective paper discussing some of the issues and choices that arise in selecting the most appropriate methods for surveillance for AMR in the environment from the perspective of a government agency that conducts routine environmental monitoring for other health-related purposes.
Environmental antimicrobial resistance: A review of biological methods. Published 26 October 2023 on gov.uk, this report reviews biological methods for the detection of environmental antimicrobial resistance.
Antimicrobial resistance surveillance strategies within wild flora and fauna of England. Published 26 October 2023 on gov.uk, this report reviews potential antimicrobial resistance surveillance strategies for wild flora and fauna.
Shellfish as bioindicator for coastal antimicrobial resistance. Published 26 October 2023 on gov.uk, this report covers development of testing approaches to assess the presence and identity of antimicrobials and resistant microorganisms in marine shellfish.
A review of approaches to monitoring and surveillance of antimicrobial resistance in bathing waters. Published 26 October 2023 on gov.uk, this report improves understanding of antimicrobial resistance in relation to human exposures via bathing waters.
Antimicrobial resistance in bioaerosols: towards a national surveillance strategy. Published 26 October 2023 on gov.uk, this report considers the decisions that must be made in designing a national surveillance strategy for antimicrobial resistance in bioaerosols.
Pilot surveillance of antimicrobial resistance in river catchments in England. Published 24 October 2024 on gov.uk, a pilot approach to detection, identification and quantification of antimicrobial resistance in three selected river catchments in England.
Development of experimental approaches for determining concentrations of antifungals that select for resistance. Published 24 October 2024 on gov.uk, methods for the determination of the lowest concentration of antifungals that can lead to a selective advantage for resistant organisms.
Determining selective concentrations for antibiotics and antifungals in natural environments. Published 24 October 2024 on gov.uk, this project determined the concentrations of specified antimicrobials at which selection for resistance may occur.
Determining concentrations of substances that influence development of antimicrobial resistance. Published 24th October 2024 on gov.uk, this report reviews the available data on concentrations at which selection for antimicrobial resistance has been reported for different antimicrobials and the approaches used to determine these concentrations. This work was also published as a peer-reviewed paper: A critical meta-analysis of predicted no effect concentrations for antimicrobial resistance selection in the environment. Published 20 August 2024 in Water Research.
Potential impact of disinfectants on antimicrobial resistance development. Published 24 October 2024 on gov.uk, this report identifies the range of disinfectants currently used in the UK and reviewed available information on their potential role in the development of antimicrobial resistance.
Risk screening and prioritisation tool for antimicrobial resistance in the environment. Published 24 October 2024 on gov.uk, a risk screening and prioritisation tool to assess antimicrobial resistance in the environment.
Cefas publications
Development and validation of a duplex RT-qPCR assay for norovirus quantification in wastewater samples. Published November 2023 this article covers development of a duplex RT-qPCR assay that enables the sensitive detection of both NoVGI and NoVGII in wastewater-derived RNA eluents, in a time and cost-effective way and may be used for surveillance to monitor public and environmental health.
Long Amplicon Nanopore Sequencing for Dual-Typing RdRp and VP1 Genes of Norovirus Genogroups I and II in Wastewater. Published on 27 March 2024 on bioRxiv, this article covers the development and optimisation of a long amplicon nanopore-based method for dual-typing the RNA-dependent RNA polymerase (RdRp) and major structural protein (VP1) regions of norovirus, with wastewater as the sample matrix.
Realising a global One Health disease surveillance approach: insights from wastewater and beyond. Published 22 June 2024 in Nature Communications, this article presents case studies centred around the recent global approach to tackle antimicrobial resistance and the current interest in wastewater testing, with the concept of 'one sample many analyses' to be further explored as the most appropriate means of initiating this endeavour.
Food-borne disease risk: biosurveillance in water networks. Published 12 September 2024 in Eurosurveillance, this meeting report details to presentations, group discussions, conclusion and recommendations of a workshops hosted by Centre for Environment, Food and Aquaculture Science (Cefas) and Bangor University at the Royal Institution, London, on 31 January 2024.
Piloting wastewater-based surveillance of norovirus in England. Published 1 October 2024 in Water Research, this study took approximately 3,200 samples of wastewater from across England, previously collected for quantification of SARS-CoV-2, and re-analysed them for the quantification of norovirus genogroup I (GI) and II (GII). Comparisons of national average norovirus concentrations in wastewater against concomitant norovirus reported case numbers showed a significant linear relationship.
VMD publications
Veterinary Antimicrobial Resistance and Sales Surveillance 2023. Published 19 November 2024 on gov.uk, reports the results from AMR surveillance pilots in dairy cattle, beef cattle and sheep.
Bangor University publications
Metagenomics unveils the role of hospitals and wastewater treatment plants on the environmental burden of antibiotic resistance genes and opportunistic pathogens. Published 10 January 2025 in Science of The Total Environment, this article cover the results from a year-long study, collecting and analysing effluent from three large hospitals, and treated and untreated wastewater from three associated community waste water treatment plants.
High-Throughput qPCR Profiling of Antimicrobial Resistance Genes and Bacterial Loads in Wastewater and Receiving Environments. Published 22 March 2025 in Environmental Pollution, this article covers the regional-based study that quantified antibiotic resistance genes (ARGs), mobile genetic elements (MGEs), and bacteria in hospital and community-derived wastewater and receiving environments, using high-throughput qPCR (HT-qPCR).
Methodology publications
Data
FSS: Scottish E.coli (locate sequencing by searching the ‘Comment’ field for ‘PATH-SAFE’ and ‘PATH-SAFE project’
Cefas: Whole Genome Sequencing Salmonella in Scottish Wastewater
University of Oxford: Campylobacter isolate collection on PubMLST
AFBI: Listeria Sequences
AFBI: Salmonella sequences (locate sequencing by searching the ‘Comment’ field for ‘PATH-SAFE’ and ‘PATH-SAFE_23_24’)
Bangor University: Metagenome from wastewater samples
Overview
The evaluation aims to identify areas for learning and improvement and the outcomes of PATH-SAFE that improve surveillance infrastructure.
There are three main objectives of the evaluation:
- design and framing: to design a table of change (ToC) for the programme, articulating the change that is intended to be achieved by PATH-SAFE - this includes outlining assumptions, external factors and managing risks associated with the programme
- formative process evaluation: to utilise bespoke evaluation methods for assessing the processes underpinning the programme, and delivering the outputs of the four workstreams to identify areas for learning and improvement
- understanding impact: to identify and prioritise outcome indicators and data sources to validate the outcomes of the ToC and develop an assessment of PATH-SAFE’s contribution
Delivery partner
RAND Europe
Outputs
- PATH-SAFE Final Evaluation Report, published on 23 June 2025
- PATH-SAFE Phase 1 Evaluation Report, published 31 October 2024
- All projects were presented at the PATH-SAFE Biosurveillance Conference on 28 and 29 February 2024 in London. Recordings, slides and posters are available on the conference webpage
- Evaluation Framework Report - Evaluation of the PATH-SAFE programme
Newsletters
For more information and updates on the PATH-SAFE programme, read the latest PATH-SAFE newsletter:
- PATH-SAFE Newsletter March 2025
- PATH-SAFE Newsletter December 2024
- PATH-SAFE newsletter September 2024
- PATH-SAFE newsletter July 2024
- PATH-SAFE newsletter March 2024
- PATH-SAFE newsletter December 2023
- PATH-SAFE newsletter September 2023
- PATH-SAFE newsletter June 2023
- PATH-SAFE newsletter March 2023
PATH-SAFE Biosurveillance Conference
On Wednesday 28 and Thursday 29 February 2024, a two-day PATH-SAFE Biosurveillance Conference was held in London. The aim of the conference was to facilitate knowledge exchange within the biosurveillance community, by showcase the biosurveillance work that has been undertaken within the PATH-SAFE programme initiatives, whilst and also creating an opportunity for people to connect and expand their professional networks.
You can access further information, including the presentation recordings, slides and posters on the PATH-SAFE Biosurveillance Conference page.
Innovation in Biosurveillance event
In November 2023, PATH-SAFE instigated and hosted a biosurveillance event that brought together over 100 key biosurveillance stakeholders from across government, academia and industry. The event actively promoted information sharing. Within facilitated workshops sessions systematic barriers were discussed, potential solutions identified, biosurveillance initiatives mapped and future opportunities explored.
Webinars
PATH-SAFE ran a series of webinar between March 2023 and January 2024 which to showcase results from the programme, as well topics relevant to programme partners.
15 March 2023 - Understanding source attribution, infection threat and level of AMR of E. coli in Scotland using whole genome sequencing
With Dr Adriana Vallejo-Trujillo (Research Fellow, Food Standards Scotland).
Please note the following resources are not accessible. If you require an accessible alternative please contact fsa.communications@food.gov.uk.
Slides available on request.
27 April 2023 - Genomic epidemiology of foodborne pathogens and antimicrobial resistance
A talk from Professor Alison Mather.
Unfortunately, due to the sensitivity of content, we are unable to share a recording or slides.
18 May 2023 - PATH-SAFE and The Food Safety Research Network Showcase
Due to a technical issue it was not possible to record this webinar. Slides available on request.
15 June 2023 - Rapid diagnostic technologies for Foodborne Pathogens
Please note the following resources are not accessible. If you require an accessible alternative, please contact fsa.communications@food.gov.uk.
Slides available on request.
13 July 2023 - First steps towards an environmental surveillance system for Antimicrobial Resistance (AMR) in England
Unfortunately, due to the sensitivity of content, we are unable to share a recording or slides.
25 September 2023 - AMR Surveillance Pilots: Dairy Cattle and Animal Feed Surveys webinar slides
Slides available on request.
17 October 2023 - PATH-SAFE Process Evaluation
Please note the following resources are not accessible. If you require an accessible alternative, please contactfsa.communications@food.gov.uk.
Slides are available on request.
9 November 2023 - Genomics of Animal and Plant Health Disease Centre II (GAP-DC2)
Unfortunately, due to the sensitivity of content, we are unable to share a recording or slides.
17 January 2024 - Genomics of antimicrobial resistant Campylobacter transmission through UK Agri-Food systems
Unfortunately, due to the sensitivity of content, we are unable to share a recording or slides.