PATH-SAFE newsletter, March 2023
The Pathogen Surveillance in Agriculture, Food and the Environment (PATH-SAFE) programme newsletter.
Welcome to the Pathogen Surveillance in Agriculture, Food and the Environment (PATH-SAFE) programme newsletter.
PATH-SAFE is a £19.2 million Shared Outcomes Fund (SOF) research programme which aims to pilot a national surveillance programme for foodborne diseases and antimicrobial resistance.
- PATH-SAFE webinar series: The programme team are starting to rollout a series of webinars showcasing results from the programme, as well as invited speakers to discuss topics relevant to programme partners. The first session, presented by Dr Adriana Vallejo-Trujillo (Research Fellow, Food Standards Scotland), will take place at 10am on 15 March 2023 titled “Understanding source attribution, infection threat and level of AMR of E. coli in Scotland using whole genome sequencing”. If you have not already received an invite but would like to attend, please contact pathsafe@food.gov.uk.
- British Science Week 10-19 March 2023: British Science Week is a ten-day celebration of science, technology, engineering, and maths. This year's theme is connections. Look out for our PATH-SAFE blog post highlighting our Communities of Interest.
Spotlight: Data Platform Consortium
We are thrilled to announce that work on the WS1a data platform development has officially begun! This project is led within the programme by FSA and will be delivered by Digital Epidemiology Services, a newly established, not-for-profit delivery arm of the Centre for Genomic Pathogen Surveillance, University of Oxford (WP1). Work has begun in earnest and consortium partners came together for a delivery kick off meeting on 20 February 2023.
An overview of the anticipated system structure and associated work packages is shown in Diagram 1 below. The data platform will utilise existing PathogenWatch infrastructure and will be hosted by CLIMB BIG DATA. The platform will allow raw sequence data to be uploaded by providers, with linkable, unique identifiers instantly generated by the system before the data is run through an assembly pipeline and genomic analysis (WP2). Visualisations and reports will be generated by the data platform and provided to end users. An assembly pipeline validation and implementation will be conducted (led by Enterobase), with interoperability improvements to the Enterobase and PubMLST systems carried out in parallel (WP3). Agreement on current best practice will be sought by community consensus (WP4).
This pilot system will use Salmonella as an exemplar pathogen and in the coming weeks, the team will look to re-engage with end users from government organisations involved in the identification and management of Salmonella outbreaks to continue to build on learnings from the discovery phase. Several subject matter experts will also be invited to participate in one of four consortium advisory groups. This community input (WP4) will allow end user reporting needs to be understood and revised iteratively. The intention is for the WS1a data platform to produce helpful visualisations and easily interpretable results that enable the rapid use of data generated by the platform.
Diagram
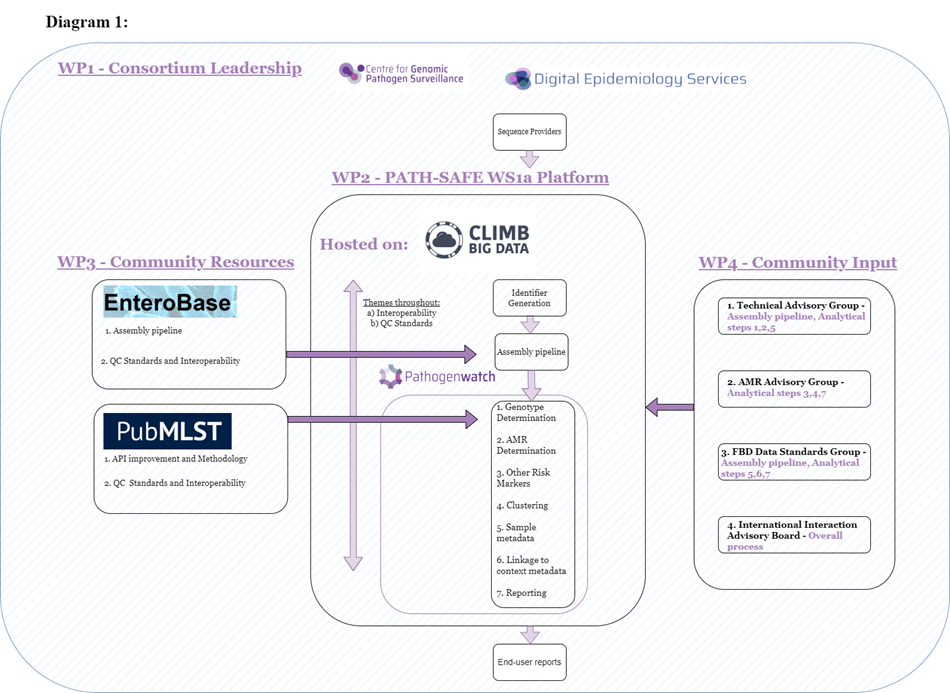
WP1 Consortium Leadership
Centre for Genomic Pathogen Surveillance and Digital Epidemiology Services.
WP2 PATH-SAFE Ws1a Platform (sequence providers):
Hosted on: Climb Big Data, themes throughout a) Interoperability b) QC Standards
- Identifier Generation
- Assembly pipeline
- Genotype Determination
- AMR Determination
- Other Risks Markers
- Clustering
- Sample metadata
- Linkage to context metadata
- Reporting
- End-user reports
WP3 Community Resources:
EnteroBase: Assembly pipeline and QC standards and Interoperability
PubMLST: API improvement and methodology and QC Standards and Interoperability.
WP4 Community Input:
- Technical Advisory Group: Assembly pipeline, analytical steps 1, 2, 5
- AMR Advisory Group: Analytical steps 3, 4, 7
- FBD Data Standards Group: Assembly pipeline, analytical; steps 5, 6, 7
- International Interaction Advisory Board – overall process
Workstream Updates
Continued progress is being made across the programme. Below are some brief updates but for more detail on each workstream please see our website.
WS1 - National foodborne disease genomic data platform
WS1a Update: The WS1a data platform development is well underway. Contracts have been signed and a kick off meeting has been held with consortium partners. Initial engagement with community advisory groups will be undertaken over the next month. Using the findings from the previous two pieces of discovery work, the system specifications for the data platform is taking shape as described above.
WS1b Update: Isolation of E. coli from shellfish, wastewater, abattoir animals and food samples continue as planned, as well as analyses:
- Additional food sampling opportunities have been identified and sampling is underway. Additional samples include positive isolates obtained from local authority food sampling, as well as 120 additional unpasteurised dairy and raw meat samples.
- Possible quality issues identified recently with wastewater samples (thought to be related to the DNA extraction process) are now fully understood. Further information on this point will be shared with the wastewater community of interest group. Volume of re-sequencing required is less than anticipated and this has begun.
- 93 E. coli isolates from shellfish are in the final stages of quality control checks before being shipped for Whole Genome Sequencing (WGS) this month.
- Engagement with Public Health Scotland and the Scottish E. coli Reference Laboratory continues; it is hoped that access to additional samples/data to bolster analyses will be possible in the coming months.
- Preparations towards producing the first analysis report by April are underway.
WS2 – New surveillance approaches
WS2a Update: In the Taw-Torridge catchment pilot, hydrodynamic model development is progressing well and wastewater, river water and shellfish sampling and analysis continue as planned, following initiation in January. Stakeholder engagement is ongoing to understand catchment activities (e.g., farming; wastewater discharges, etc) that may contribute to pathogen loadings in specific areas of the study catchments. Wastewater sampling and analysis have continued at the North Wales catchment pilot site, with an additional autosampler to be set up in March. Detection of norovirus in UKHSA COVID-19 archived wastewater samples ongoing at the Environment Agency, with associated WGS to begin next month at Cefas. Arrangements for Scottish wastewater Salmonella pilot study are being finalised with bacteriology work to start in March.
WS2b Update: Sequencing of the raw meat E. coli isolate is drawing to a close, with two batches (totalling 163 isolates) complete and the third batch (20 isolates) to be sequenced in March, followed by the start of bioinformatic analysis in April. Continued engagement with industry has resulted in the successful initiation of sample collection on the sheep survey project, alongside sample collection and analysis continuing as planned on the raw milk project and animal feed project (raw ingredients and finished feed). Piloting of methods and survey design are progressing well for the cattle survey project. Continued engagement with UK Pet Food to scope out potential for future collaboration with raw pet food plans.
WS2c Update: Wastewater sampling across 5 sites in Belfast progressing well. Additional AMR detection modules for MALDI-ToF installed. Surfaces for stability of SPED (silica nanoparticles with encapsulated DNA) testing identified and SPED stability testing will continue in March to confirm stability under environmental conditions and on different surfaces, to understand potential real-life conditions. Protocols for whole genome sequencing established and ongoing identification of appropriate databases for post sequencing AMR analysis that offer comprehensive tools for the rigorous examination of resistance genes, their products, and associated phenotypes e.g., ABRicate, ResFinder and CARD.
WS2d Update: This project will investigate source and transmission dynamics of food borne disease (FBD) caused by Campylobacter, including investigation of the spread of antimicrobial resistance (AMR) through Agri-Food systems. The project will do so using existing, comprehensive isolate collections which are ready to be sequence typed. Contracting has just concluded and initial activities will include engaging with stakeholders to arrange sample shipments, completing DNA extractions, and making the relevant preparations for sample sequencing.
WS2e Update: 100 archived Salmonella isolates selected for sequencing, representing a spread of serovars, sampling years and sources from within the available isolates. The culture process will be initiated in February and then the isolates will be whole genome sequenced.
WS3 – Rapid, in-field diagnostic technologies
WS3a Update: Screening of the titles and abstracts of papers paper identified during literature search is ongoing. These will be used to establish an initial database of technologies identified, which will be expanded as full texts are reviewed. Technology readiness level (TRL) definitions are being finalised ahead of delivery at the end of February. Outreach to stakeholders and potential end users has generated a good range of responses, a few additional targeted outreaches will be made to further broaden representation ahead of workshops in March and April.
WS3b Update: 20/30 Labs have been making sustained progress on the proof-of-concept LAMP (versus qPCR) to identify 7 target pathogen - Salmonella spp., Listeria monocytogenes, Norovirus, adenovirus, astrovirus, rotavirus and sapovirus. Progress has also been made on the next stage of optimising the LAMP assay for the 7 target pathogens, with work to transition from a buffer matrix to wastewater at the end of February.
WS4 - Environment AMR surveillance system pilot
WS4 Update: Sampling of surface water at the pilot river catchments (phases 1-3) is complete. Analysis (MIC testing and high throughput detection and quantification of Antibiotic Resistance with Resistomap) continues. The following other R&D projects are in progress:
- ‘AMR shellfish’ project with Cefas
- ‘Bioaerosol’ project with WSP, Cranfield university and University of Exeter
- 'Disinfectant' project with University of Exeter, UKCEH and Cardiff University
- ‘Bathing Waters’ project work undertaken by Atkins and UKCEH
- Three projects reviewing and exploring different aspects in relation to minimum selection concentrations for antifungals and antibiotics with University of Exeter and UKCEH.
The following R&D projects are in the final stages/have concluded:
- Antifungals in the terrestrial environment; a paper has recently been published from this work
- Wild flora and fauna project (report under review)
- Discovery work for a future AMR One Health Surveillance System (supported by Deloitte) is ongoing. Defra contribution to this has recently concluded with the production of an options paper which sets out technical needs, costings, and anticipated challenges.
- Development of the exemplar AMR Environmental Surveillance System (supported by Deloitte) continues. The Beta phase began in February 2023 and is expected to conclude by end of March 2023.
Meet the team
Each quarter we will spotlight people working across the programme. In this issue we are focussing on colleagues who are leading the Data Platform Consortium in WS1a.
The consortium is led by colleagues from the Centre for Genomic Pathogen Surveillance (CGPS) and Digital Epidemiology Services (DES) at the University of Oxford.

Professor David Aanensen, Director CGPS, Professor of Genomic Surveillance & PATH-SAFE Data Fellow
David brings together a unique blend of expertise in data modelling, software development, epidemiology, bioinformatics and machine learning and genomic technology. Within CGPS, David and team focus on data flow and the use of genome sequencing for surveillance of microbial pathogens through a combination of web application and software engineering, methods development and large-scale structured pathogen surveys and sequencing of microbes with delivery of information for decision making. David is also Director of the NIHR Global Health Research Unit on Genomic Surveillance of AMR which has delivered sustainable genomic and data capacity for AMR surveillance with partners in India, Colombia, Nigeria, and The Philippines. This work has helped shape policy within the country and upwards within white papers for the World Health Organisation (WHO). David is the PATH-SAFE Data Fellow and has been instrumental in driving forward innovation and leading on the translation of complex strategic requirements into achievable deliverables.

Georgina Haines-Woodhouse, Digital Epidemiologist & Data Scientist
Utilising her background in molecular biology, data science and epidemiology, Georgina has come on board the PATH-SAFE programme as lead digital epidemiologist and data scientist. Previously, Georgina worked for the Global Research on Antimicrobial Resistance (GRAM) project at the University of Oxford with a focus on modelling the prevalence of resistance of Staphylococcus aureus and Streptococcus pneumoniae as well as possible factors that drive AMR. She has a Master of Research (MRes) in Ecology, Evolution and Development and a BSc in Biology from Oxford Brookes University.

Corin Yeats, Genomic Software Engineer
Corin is an experienced developer of genomics research software, including a decade at CGPS, and is an author of fifty peer-reviewed publications. After completing a PhD in microbial genomics and the evolution of protein function within the Pfam team at the Welcome Sanger Institute, he joined the CATH-Gene3D team at University College London as a post-doctoral researcher into the structural classification of protein domains and relation to function. This post included multiple collaborative projects involving national and international consortia, including the pan-European BioSapiens network. After moving to the CGPS, he has subsequently focused on the delivery of genomics software to the public health community and has been involved in writing or adapting many of the tools that underlie the Pathogenwatch system.

Angela Blanton, Programme Manager
Angela is the lead programme manager for WS1a. She liaises with both stakeholders and site personnel, coordinating efforts within the CGPS/DES team to ensure timely reporting and achievement of deliverables. Her extensive experience includes several years in public health as programme manager for data management improvement projects for Florida DoH, Bureau of Public Health Laboratories, as well as several years’ experience as a senior clinical data analyst and team lead for multiple oncology studies and a program analyst supporting the U.S. government. She holds a BSPH from the Gillings School of Global Public Health, University of North Carolina at Chapel Hill.

Diana Connor, Senior Programme Manager, CGPS
Diana is the senior programme manager for CGPS having oversight of DES and CGPS’s involvement in WS1a. Diana obtained her MPH of infectious Disease and Microbiology at the University of Pittsburgh, USA and was the co-coordinator of a public health computational modelling group which conducted vaccine supply chain research in various countries across the globe. Most recently, she was the consortium coordinator in Copenhagen, Denmark, for an EU wide One Health project through the OneHeath European Joint Programme. Diana has co-authored over 25 peer reviewed journal articles focusing on infectious disease and public health.

Steve Karp, Operations
Steve’s role in PATH-SAFE is to ensure operational delivery is kept on track, as well as supporting stakeholder management. He joined CGPS to assist in scaling up capabilities and has helped to establish Digital Epidemiology Services (DES) to implement those capabilities sustainably across the globe. He has extensive expertise in all aspects of alliance, program, and operations management in the life sciences. He has directed development and tech transfer programs in a variety of technologies. He has run various collaborative research and development programs working with government, non-profits, and private industry. He attended Harvard College.
Further information
For any questions or feedback please contact the team at pathsafe@food.gov.uk.
To sign up to the SERD newsletter which contains PATH-SAFE news and link to our blog please visit Food Standards Agency UK (govdelivery.com).
Revision log
Published: 27 February 2023
Last updated: 28 February 2024