Pathogen Surveillance in Agriculture, Food and Environment (PATH-SAFE) Programme
The PATH-SAFE programme piloted the development of a national surveillance network, using the latest DNA-sequencing technology and environmental sampling, to improve the detection and tracking of foodborne human pathogens and associated AMR through the whole agri-food system from farm-to-fork.
PATH-SAFE was a 4-year, cross government and UK wide programme, led by the Food Standards Agency (FSA) that utilised £24m of funding from the HMT Shared Outcomes Fund (SOF) and match funding from a range of government and academic delivery partners. The programme piloted the development of a national surveillance network, using the latest DNA-sequencing technology and environmental sampling, to improve the detection and tracking of foodborne human pathogens and associated AMR through the whole agri-food system from farm-to-fork.
PATH-SAFE ended in March 2025, however this page will continue to be updated with links to programme outputs, and will transition to include information about follow on biosurveillance work in due course.
In October 2024, a commentary summarising the program's achievements, and consideration on how they could be taken forward beyond the lifetime of the program, was published in Future Microbiology (October 2024): Coordinated surveillance of foodborne pathogens and antimicrobial resistance: insights from the PATH-SAFE pilot.
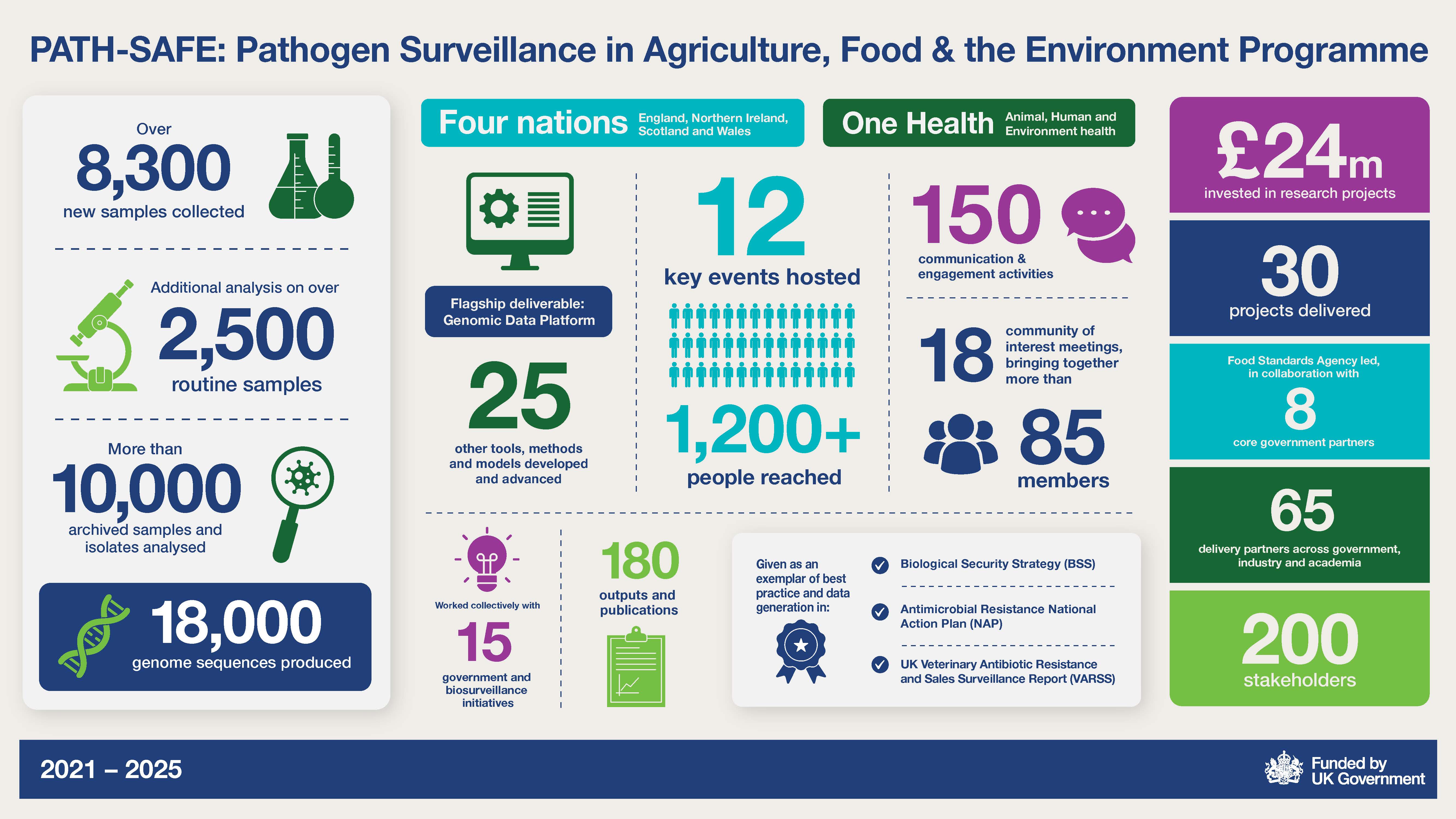
£24 million invested in research projects. 30 projects delivered. Food Standards Agency led, in collaboration with 8 core government partners. 65 delivery partners across government, industry and academia. 200 stakeholders. 2021 to 2025: Over 8300 new samples collected. Additional analysis on over 2500 routine samples. More then 10000 archived samples and isolates analysed, 18000 genome sequences produced. Four nations - England, Northern Ireland, Scotland and Wales. One Health - Animal, Human and Environment health. Flagship deliverable: genomic data platform 25 other tools, methods and models developed and advanced. 12 key events hosted. 1200 people reached. 150 communication and engagement activities. 18 community of interest meetings, bringing together more than 85 members. Worked collectively with 15 government and biosurveillance initiatives. 180 outputs and publications. Given as an exemplar of best practice and data generation in Biological Security Strategy (BSS), Antimicrobial Resistance National Action Plan (NAP), UK Veterinary Antibiotic Resistance and Sales Surveillance Report (VARSS)
Programme background
Foodborne disease (FBD) is a major public health risk with 2.4 million individual illnesses and more than 16,000 hospitalisations per year. Most human disease is caused by a handful of bacteria which enter the food chain from farmed animals or the environment.
In addition to FBD, the agri-food supply chain also poses a risk for the transmission of antimicrobial resistance (AMR) through food, animals, humans, and water. Whilst the UK has made progress in reducing its use of antibiotics in humans and animals in the last five years, drug-resistant bloodstream infections in humans have increased by 32% from 2015 to 2019. The rise and spread of AMR is creating a new generation of ‘superbugs’ that cannot be treated with existing medicines.
For these reasons, government departments already undertake surveillance activities by analysing samples from food, livestock, and humans. Advances in technology and data management offer the opportunity to create a step change in surveillance, to protect public health. The PATH-SAFE programme established a new data platform that allows for the analysis, storage and sharing of pathogen sequence and source data, collected from multiple locations across the UK by government departments and public organisations. This single system will enable rapid identification and tracking of FBD and AMR. This will improve public health and minimise the economic and environmental impact of outbreaks.
Aims of the programme
- To pilot a better national surveillance system for the monitoring and tracking of FBD and AMR in the environment and agri-food system
- To bring together and build on existing initiatives across the UK and to understand what the end-user needs to improve how they work in this space
- To provide better data to identify the prevalence, source and pathways of FBD and AMR, helping to prevent spread by enhanced targeting of interventions
Benefits of the programme
- Generation of information and knowledge that support enhanced identification of FBD and AMR sources
- Providing improved user access to relevant data so they can make more informed, evidence-based decisions
- Data that underpins the prevention and prediction of FBD and AMR spread by supporting improved, cost-effective targeting of interventions, providing economic savings both for government and industry
- Improved surveillance system that supports better identify and quicker reaction to emergent diseases (or diseases of increasing concern).
- Potential to reduce the number and cost of FBDs and AMR, lower commercial losses, strengthen UK Science Excellence, and enhance the UK food sector’s reputation
Programme partners
The PATH-SAFE programme was led by the FSA, supported by several core partners:
- Food Standards Scotland (FSS)
- Department of Environment, Food and Rural Affairs (Defra)
- Animal and Plant Health Agency (APHA)
- Centre for Environment, Fisheries and Aquaculture Science (Cefas)
- Environment Agency (EA)
- Veterinary Medicines Directorate (VMD)
- Department of Health and Social Care (DHSC)
- UK Health Security Agency (UKHSA)
The programme was delivered through a one-health, four-nation collaborative approach, both at the delivery level and at the strategic level.
More than 65 organisations, from across government, academia and industry, have been involved in delivery and advisory roles. These have included:

Department for Environment, Food and Rural Affairs, Animal & Plant Health Agency, UK Health Security Agency, Centre for Environment, Fisheries and Aquaculture Science, Veterinary Medicines Directorate, Department of Health and Social Care, Food Standards Scotland, Scottish Environment Protection Agency, Scotland's Rural College, Public Health Scotland, Food Standards Agency, Environment Agency, Northern Ireland Environment Agency, NHS Lothian, Department of Heath (NI), Agriculture, Environment and Rural Affairs (NI), Department of Infrastructure (NI), Welsh Government, HSC Public Health Agency, Northern Ireland Water, Agri-food and biosciences institute, Public Health Wales, Welsh Water, South West Water, Ribble Rivers Trust, Biosphere North Devon, Virosafety, resistomap, Quadram Institute, Moredun Research Institute, EnteroBase, Queen's University Belfast, University of Lincoln, Cardiff University, Climb big data, The University of Edinburgh, University of Exeter, Cranfield University, Bangor University, The University of Warwick, University of Oxford, University of Birmingham, University of Worcester, UK Pet Food, Agricultural Industries Confederation, Arla, Fera, UK Centre for Ecology and Hydrology, Centre for Genomic Pathogen Surveillance, Digital Epidemiology Services Ltd, Aecom, Arup, Capita, Deloitte, National Milk Laboratories, MicrobesNG, RAND Europe, NHS Greater Glasgow and Clyde, The City of Edinburgh Council, Glasgow City Council, Met Office, Channel Costal Observatory, PubMLST, Pathogenwatch, Nijhuis
Funding source and timeline
PATH-SAFE was funded via the Treasury Shared Outcomes Fund (SOF).
Spending Round 2019 announced £200 million for the Shared Outcomes Fund to fund pilot projects to test innovative ways of working across the public sector. The first round of the SOF funded a wide range of projects to be run in 2020-21 and 2022-23.
Spending Review 2020 announced a further £200 million was to be made available for a second round of the Shared Outcomes Fund between 2021-22 and 2023-24.
In 2021 the programme successfully secured £19.2m in phase 2 of the SOF, to run a three-year programme concluding in March 2024.
At the start of 2024, PATH-SAFE successfully secured an additional £4.7m of funding to continue for an additional year (April 2024 to March 2025), with HMT SOF providing £2.2m of the continuation funding, leveraged by £2.5m of match funding from PATH-SAFE core government partner departments and academic delivery partners.
Programme themes
The programme was organised thematically for governance, oversight, reporting and coordination purposes.
Further information about each of the themes, including overviews, delivery partners, status, key successes and outputs can be found on the following pages:
- National foodborne disease genomic data platform
- Onsite Diagnostics
- Novel Foodborne Pathogen, and associated antimicrobial resistance, knowledge and surveillance approaches
- Programme Evaluation
The programme developed new systems and tools, explored novel surveillance approaches and generated knowledge, to improve the detection and tracking of foodborne human pathogens and antimicrobial resistance, through the whole agri-food system from farm-to-fork.

Alongside the technical advances the programme has driven forward, PATH-SAFE has also established and promoted a collaborative, four nation, One Health approach to surveillance, which has been referenced as an exemplar within initiatives such as the Biological Surveillance Strategy and the UK AMR National Action plan (noted within as a key achievement).
The programme has influenced, informed, and supported programmes, strategies and initiatives across government and beyond, including:
- Biothreats Radar and National Biosurveillance Network (NBN)
- UK Microbial Forensics Consortium (UKMFC)
- Genomics of Animal and Plant Disease Centre Phase 2 (GAP-DC2)
- AMR National Action plan (NAP) 2019-24 and 2024-29
- UKRI Tackling Infections Strategic Theme
- Government Office for Technology Transfer (GOTT)
- Microbiology Society – Knocking Out AMR initiative
- ONS Integrated Data Service (IDS) Programme
- Food Safety Research Network (FSRN)
Events and communications
Throughout the programme, a range of events and communication activities have been undertaken including a webinar series, a conference and publication of quarterly newsletters.
Further information on these events and activities can be found on the following pages:
Get in touch
If you would like to get in touch with the PATH-safe team, you can email them at pathsafe@food.gov.uk
Revision log
Published: 22 August 2022
Last updated: 10 April 2025