Critical review of AMR risks arising as a consequence of using biocides and certain heavy metals in food animal production
Antimicrobial resistance (AMR) is the resistance of a microorganism to an antimicrobial agent (a substance that kills or stops the growth of microorganisms) that was originally effective for treatment of infections caused by it. As a result standard antimicrobial drug treatments may become ineffective, lead to infections persisting, increasing the risk of spread to others, and negative clinical outcomes. AMR is a major public health issue worldwide and it is estimated that unless action is taken to tackle AMR, the global impact of AMR could be 10 million deaths annually from drug-resistant infections by 2050 and cost up to US $100 trillion in terms of cumulative lost global production (O’Neill, 2016). Addressing the public health threat posed by AMR is a national strategic priority for the UK and led to the Government publishing both a 20-year vision of AMR and a 5-year (2019 to 2024) AMR National Action Plan (NAP), which sets out actions to slow the development and spread of AMR. Intensive food animal production plays an important role in the development and spread of AMR and is one of many routes by which consumers can be exposed to antimicrobial-resistant bacteria. This review was carried out to help increase our understanding of whether, and to what extent, the use of biocides (disinfectants and sanitisers) and heavy metals (used in feed and other uses) in animal production leads to the development and spread of AMR within the food chain (a subject highlighted in the NAP). Whether this could potentially lead to greater consumer exposure to antimicrobial-resistant bacteria present in our food, either directly through consumption of foods derived from animals that have undergone treatment (for example from the use of heavy metals in animal feed) or indirectly (for example from exposure of crops to contaminated soil or ground water) is not known.
Focused searching of three literature databases (Web of Science, Scopus, and MEDLINE) was undertaken, supplemented by additional records identified through other sources. Due to the range of publications identified and different laboratory methodologies used in these studies no statistical analysis was possible, so instead, a narrative approach was taken to their review and to the review of supplementary materials.
We conclude that there is published evidence that the release of chemicals like biocides (in particular disinfectants) and/or heavy metals from food animal production have the potential to contribute to the selection, emergence, and spread of AMR (as bacteria or genes) that could be acquired by consumers, and that this could present a potential risk to the consumer as a result. The published evidence is sparse and there are significant knowledge gaps (as detailed in this report). Currently there are insufficient data for a comprehensive and quantitative assessment of risk, and a need for focussed in-field studies (as detailed in this report) to be carried out to fill these knowledge gaps and confirm whether there is an actual risk.
Antimicrobial resistance (AMR) is the resistance of a microorganism to an antimicrobial drug that was originally effective for treatment of infections caused by it, so that standard treatments become ineffective, and infections persist, increasing the risk of spread to others and negative clinical outcomes. AMR is a complex issue driven by a variety of interconnected factors enabling microorganisms to withstand the killing or microstatic effects of antimicrobial treatments, such as antibiotics, antifungals, biocides, and preservatives. Microorganisms may be inherently resistant to such treatments or can change and adapt to overcome the effects of such treatments. Microorganisms can acquire AMR due to mutation or through horizontal gene transfer (HGT) via several mechanisms. The widespread use of antimicrobials in clinical practice and especially in intensive food animal production is known to result in selection and dissemination of AMR in microorganisms. AMR and antimicrobial resistance genes (ARGs) are a major public health issue worldwide and it is estimated that unless action is taken to tackle AMR, the global impact of AMR could be 10 million deaths annually by 2050 and an associated cost of up to US $100 trillion in terms of cumulative lost global production (O’Neill, 2016).
Biocides are active chemical molecules (agents) that control the growth of, or kill, bacteria and other microorganisms. Disinfectants and sanitisers are forms of biocides. Biocides are used for a number of reasons in animal production including, the cleaning and disinfecting of buildings and equipment as well as decontaminating ponds and equipment in fish farming. The primary use of heavy metals, such as copper and zinc, in food animal production are as nutritional additives in animal feed but they may be used in livestock footbaths and to treat skin problems such as dermatitis. Some biocides and heavy metals used in animal husbandry can persist and concentrate in the environment, remaining stable for prolonged periods. It is a concern that bacteria can exhibit tolerance to these chemical and metal elements and that the genes encoding for these phenotypes can be located on mobile elements that may contain one or more AMR- encoding genes, thereby co-selecting for AMR (as highlighted in the NAP).
This study was undertaken to critically review the available scientific literature for assessing whether, and to what extent, the single or combined effects of using widely available biocides for sanitation and heavy metals in animal feed and other uses (such as therapeutically and to treat infections) during food animal production leads to the development and spread of AMR within the food chain. Whether this usage could potentially lead to greater consumer exposure to antimicrobial-resistant bacteria from food, either directly through consumption of foods derived from animals that have undergone treatment (for example from the use of heavy metals in animal feed) or indirectly (for example from exposure of crops to contaminated soil or ground water) is also discussed.
The review question was defined as:
“Do biocides and/or heavy metals used in food animal production have an impact on the development of AMR in the food chain?”
A systematic review approach was taken to the literature search. The review adopted a comprehensive search strategy considering all available evidence in the public domain, including peer-reviewed articles, grey literature (for example, government and industry reports), relevant government reports (for example, FSA published studies, ACMSF reports), European and International literature (for example, the EFSA Scientific Opinions, WHO reports) up to February 2023. Three scientific literature databases (Web of Science, Scopus, and MEDLINE) were searched for relevant publications, supplemented by focused Google and Google Scholar searches, searching within relevant publications, and through contact with authors. A total of 3,434 of publications were identified, which were reduced to 550 after screening the titles and abstracts. This total was further reduced to 148, from which some data were extracted after appraising the full publications. Due to the range of publications identified, the paucity of specific published studies, and different laboratory methodologies used in these studies, a narrative approach was taken to their review and to the review of supplementary materials.
The review was structured and aimed at addressing the following key questions (terms of reference):
- Is there evidence in the literature to show that biocides and/or heavy metals used in food animal production have an impact on the development of AMR?
- How long are biocides and/or heavy metals (used in food animal production) able to persist in animal production environments, and how does this impact on the development of AMR and associated risks?
- What evidence from the literature is there that biocide and/or heavy metal-associated AMR enters the food chain through products of animal origin or as a result of crop contamination?
- Is there a potential risk to the consumer from AMR acquired through the use of biocides and/or heavy metals in food animal production?
On reviewing the published evidence our conclusions regarding these four key questions are that:
- We have found that there is some evidence that both biocides and heavy metals used in food animal production may have an impact on the development of AMR, and either resulting in reduced susceptibility to drugs or clinically significant resistance. There is more compelling evidence regarding the use of heavy metals than there is on the use of biocides.
- We have found that there is compelling evidence that heavy metals will persist, accumulate, and may impact on the development of AMR in animal production environments for many years. There is less evidence on the persistence and impact of biocides. There is some evidence that while many biocides will rapidly break-down in the environment, some, such as quaternary ammonium compounds (QACs), may persist. However, there are little data on how long and at what concentration such agents may persist in animal production environments, and what the impact on AMR may be.
- We have not found any clear evidence of biocide and/or heavy metal associated AMR entering the food chain, through products of animal origin or as a result of crop contamination due to their use in food animal production. Published studies that have demonstrated an association between biocide and/or heavy metal use and increased AMR/reduced susceptibility risk in live animals, manure, slurry, or soil have not looked beyond these points at how this use may impact on AMR risk in food. Although there is evidence of the co-carriage of biocide and heavy metal resistance genes and ARGs in retail meats.
- We have found that there is evidence that AMR in food is a risk, and that food animal production has an impact on AMR risk. However, while there is certainly a theoretical risk, we have found no published evidence that has specifically demonstrated that the use of biocides and/or heavy metals in food animal production increases the risk of the consumer acquiring AMR or has indeed quantified that risk. Currently there does not appear to be sufficient evidence to carry out such an assessment of risk.
A central question was whether the release of chemicals like biocides (in particular disinfectants) and/or heavy metals from food animal production has the potential to create local concentrations where AMR can emerge and spread (as bacteria or genes) and whether this presents a potential risk to the consumer as a result. In our opinion there does appear to be sufficient evidence that this is possible and that there is a potential risk to the consumer. However, currently there does not appear to be sufficient data to carry out an assessment of risk, and there is a clear need for focussed in-field studies (as detailed in this report) to be carried out to fill this evidence gap and provide the data required to assess this risk.
AMR is the resistance of a microorganism to an antimicrobial agent (a substance that kills or stops the growth of microorganisms) that was originally effective for treatment of infections caused by it, so that standard treatments become ineffective, and infections persist, increasing the risk of spread to others. In the context of clinical bacterial infections, resistance is most often defined based on likely clinical efficacy of an antimicrobial agent/bacteria combination; however clinical breakpoints (discriminatory antimicrobial concentrations used in the interpretation of results of susceptibility testing to define isolates as susceptible, intermediate, or resistant) are not available for all antimicrobial agent/bacteria combinations. This is particularly the case for agents such as biocides and heavy metals, where no internationally accepted breakpoints exist to define resistance. However, a further way of monitoring the development of AMR and any reduction in susceptibility or increase in tolerance is to use epidemiological cut-offs, which enables bacteria with reduced susceptibility to an antimicrobial agent to be distinguished from the wild type population and innate susceptibility. Within the literature, resistance is not always clearly defined, especially concerning studies investigating susceptibility/tolerance to biocides and heavy metals. For the purposes of this review the terms tolerance and reduced susceptibility are used when describing biocide and/or heavy metal “resistance”.
AMR is a complex issue driven by a variety of interconnected factors enabling microorganisms to withstand the killing or static effects of antimicrobial agents, such as antibiotics, antifungals, disinfectants, and preservatives. The widespread use of antimicrobial agents in all contexts is known to result in selection for AMR in microorganisms (O’Neill, 2016). There is also evidence that biocidal agents and/or heavy metals may, in some contexts, co-select for AMR in microorganisms (the focus of this review).
AMR and ARGs are a major public health issue worldwide and it is estimated that unless action is taken now to tackle AMR the global impact of AMR could be 10 million deaths annually from drug-resistant infections by 2050, costing up to US $100 trillion in health costs and cumulative lost economic output (O’Neill, 2016). Resistance to bacterial infections can make infections caused by these organisms difficult to treat and cause illness to persist, with recognised extra costs and increased morbidity and mortality (Likotrafiti et al., 2018).
Addressing the public health threat posed by AMR is a national strategic priority for the UK and led to the Government publishing both a 20-year vision of AMR and a 5-year (2019 to 2024) AMR National Action Plan (NAP) which sets out actions to slow the development and spread of AMR with a focus on antimicrobials. The NAP has adopted an integrated ‘One-Health’ approach which spans people, animals, agriculture, and the environment and calls for activities to “identify and assess the sources, pathways, and exposure risks” of AMR. The FSA have, and are continuing, to contribute to delivery of the 5-year NAP through furthering understanding of the role of the food chain and AMR, conserving the effectiveness of current treatments through the adoption of good hygiene practices, and encouraging the food industry to reduce usage of antimicrobials where possible. ARGs that result in resistance to what are termed ‘Critically Important Antimicrobial’s’ (CIAs) by the World Health Organisation (WHO) are of particular concern to the FSA.
AMR may be intrinsic or acquired by transfer mechanisms (Verraes et al., 2013). Transfer mechanisms include vertical gene transfer, acquired because of mutation (for example, genomic point mutations) [which in turn is passed on vertically], or the acquisition of ARGs within the same species or between different bacterial species by horizontal gene transfer [HGT] (Verraes et al., 2013; Munita & Arias, 2016). Bacteria may be resistant to just one antimicrobial agent or to several different agents (multi-resistant or multi-drug resistant (MDR) defined as resistance of a bacterial isolate to three or more classes of antimicrobials), with cross-resistance dependent on which ARGs and other mechanisms of resistance are present (such as, enzymatic, permeability barriers, and efflux pumps).
The transmission of antimicrobial-resistant microorganisms and ARGs to food and within the food chain is complex. Food can be contaminated with antimicrobial-resistant bacteria and/or ARGs in several ways (Verraes et al., 2013; Food Standards Agency, 2016) including (but not exclusively):
- Through contamination with antimicrobial-resistant bacteria in the environment.
- Through the presence of antimicrobial-resistant bacteria in food animals treated by antimicrobials during agricultural production.
- The possible presence of ARGs in bacteria that are intentionally added during the processing of food (starter cultures, probiotics, bio-conserving microorganisms, and bacteriophages).
- Through cross-contamination with antimicrobial-resistant bacteria and ARGs during food processing
Biocides
A biocide is defined as an active chemical molecule that controls the growth of, or kills, bacteria and other microorganisms in a biocidal product (SCENIHR, 2009; Wales & Davies, 2015; VKM, 2016). Biocidal substances act in different ways and sometimes several biocides are combined within a single product to increase the overall efficacy (VKM, 2016). The mechanisms of action and resistance/reduced susceptibility to a wide range of biocides on bacteria have been reviewed and described by McDonnell & Russell (1999), Ortega Morente et al. (2013), and Geueke (2014), amongst others. Many biocides act by effecting the plasma membrane of bacteria, because of which Gram-negative bacteria are generally less susceptible to many biocides than are Gram-positive bacteria (Denyer & Maillard, 2002; Wales & Davies, 2015).
Biocides are classified into different groups according to their application categories (Table 1). Biocides used in food animal production operations mainly act as disinfectants, sanitising agents, or antiseptics (SCENIHR, 2009; VKM, 2016; Donaghy et al., 2019). Examples of use include: the cleaning and disinfecting of buildings and equipment as well as decontaminating ponds and equipment in fish farming; in footbaths for operators outside animal housing; in livestock footbaths to treat and prevent the spread foot infections such as digital dermatitis; to clean udders of animals used for milk production; and for preserving specific products such as eggs or semen (SCENIHR, 2009; Wales & Davies, 2015; Donaghy et al., 2019; VKM, 2016). They may be used in anti-fouling paints used in aquaculture to reduce the growth of attached organisms on fish cages and nets (Burridge et al., 2010; Guardiola et al., 2012). Biocides are generally not used within body tissues (though some such as organic acids and essential oils (EOs) are added to animal feed and water as antimicrobial controls).
Table 1: Examples of biocidal products on the basis of chemical group (McDonnell & Russell, 1999; SCENIHR, 2009; VKM, 2016), not all example compounds listed may be permitted in the UK.
| Chemical group | Example products/compounds | Examples of use |
|---|---|---|
| Antimicrobial dyes | Acridines, triphenylmethane dyes, quinones | Disinfection of equipment in fish farming. |
| Aldehydes | Glutaraldehyde, formaldehyde, other aldehydes | Disinfection of equipment and environments in land farming and aquaculture |
| Alcohols | Ethyl alcohol (ethanol), methyl alcohol (methanol), other alcohols | Antiseptics and disinfection agents. |
| Biguanides | Chlorhexidine | General purpose disinfectant/antiseptic for cleansing wounds, skin, instruments, and equipment. Including as a dairy teat disinfectant. |
| Chlorine compounds | Sodium hypochlorite (active agent in bleach), chlorine dioxide, electrolysed water. | Widely used for both antiseptic and disinfectant purposes in drinking water, wastewater, and in fish farms. |
| Essential oils (EOs), plant compounds, and extracts | Menthol, tea tree oil, cinnamon oil, oregano oil, thyme | Disinfection, decontamination, including use in animal feed and water as antimicrobial controls. |
| Iodine-releasing agents | Free iodine, iodophors | Used in teat dips for the prevention and control of mastitis in cattle. |
| Organic and inorganic acids: esters and salts | Acetic acid (ethanoic acid), citric acid, lactic acid. | Disinfection, decontamination, including use in animal feed and water as antimicrobial controls. |
| Peroxygens | Hydrogen peroxide, peracetic acid, ozone. | Disinfection, decontamination, and sterilisation, including the treatment of waste water and foods. |
| Phenols | Creosols, non-coal tar phenols, halophenols, nitrophenols, bisphenols. | Antiseptics and disinfection agents. |
| Quaternary ammonium compounds (QACs) | Benzalkonium chloride, cetrimide (alkyl trimethyl ammonium bromide, a mixture of three QACs). | General disinfection in the food industry. |
Biocidal products are regulated as they have the potential to cause harm to human health and/or the environment. Biocidal products are controlled in Great Britain (England, Scotland, and Wales) under the GB Biocidal Products Regulation (GB BPR) and in Northern Ireland under the EU Biocidal Products Regulation (EU BPR). A list of UK- authorised biocidal products is provided by the Health and Safety Executive (HSE).
This review has focused on the impact of only those biocides used in food animal production. For this reason, triclosan (5-chloro-2(2,4-dichlorophenoxy)phenol) was not considered, as this product was used almost exclusively in human related products, such as handwashes, toothpastes and other personal care products as well as being incorporated into other consumer products such as clothes. Due to health concerns and potential impact on the environment, it has been banned within the EU 27 and also in the USA. While increased AMR to this product is often discussed in the literature, it is of limited relevance to food animal production (Davies & Wales, 2019).
Heavy metals
Heavy metals are naturally occurring elements that have a high atomic weight and a density that is at least 5 times greater than that of water (Tchounwou et al., 2012). Some heavy metals (such as cobalt, copper, iron, manganese, molybdenum, selenium, and zinc) are essential in the diet of living things to maintain various physiological functions and are usually added as nutritional supplements in animal feed (Hejna et al., 2018). They have antimicrobial properties and may be used for this purpose in food animal production. In feed and as supplements they improve growth and prevent diseases via these antimicrobial properties by acting on the gut microbiota to reduce loss of nutrients and suppress gut bacteria, including pathogens (Li et al., 2022a). They may also be used as antimicrobials, The antimicrobial modes/mechanisms of action of heavy metals on bacteria /microbes have been reviewed by Lemire et al. (2013). Different metals cause discrete and distinct types of injuries to microbial cells as a result of oxidative stress, protein dysfunction, or membrane damage (Lemire et al., 2013). Due to the presence of toxic heavy metals in the general environment, many bacteria have evolved mechanisms of metal resistance (Vats et al., 2021). As discussed further, these mechanisms of resistance/tolerance may provide resistance/tolerance to other antimicrobials leading to co-selection for AMR.
Copper and zinc are used in the pig and poultry sectors as in-feed growth promotors and for enteric disease control (Wales & Davies, 2015). Zinc is used in aquaculture as a supplement in feed (Burridge et al., 2010; Yu et al., 2021). Heavy metals are often used in higher concentrations than needed to ensure adequate nutrition (Medardus et al., 2014; Yu et al., 2017). A survey of livestock feeds in England and Wales in 1999 reported concentrations of zinc and copper of 150–2,920 ppm and 18–217 ppm, respectively, in pig feeds and 28 to 4,030 ppm and 5 to 234 ppm, respectively, in poultry feeds (Nicholson et al., 1999). Since the bioavailability of metals in feed is usually quite low, unabsorbed heavy metals are excreted in faeces and may accumulate in soil, water, and sediments from agricultural practices. One study in the USA found 90% of in-feed copper and zinc fed to pigs was shed in faeces (Medardus et al., 2014). Although recently introduced new sources or forms of these metals with higher bioavailability allows for substantial reduction of dietary inclusion rates (Dębski, 2016). The total amounts and concentrations used of copper and zinc in feed may differ among countries, due to restrictions imposed by national legislation.
In two opinions, the EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP) recommended that zinc and copper contents be cut (EFSA FEEDAP Panel, 2014, 2016). They suggested that this would reduce residues of both of these metals in manure by 20%. Recommendations for zinc were based on animal requirements, while the opinion on copper in feed also considered its impact on AMR. These recommendations have since been enacted in EU legislation. Permitted maximum zinc contents animal feed in the EU (Regulation 2016/1095) are: 180 mg zinc/kg for salmonids and in milk replacers for calves; 150 mg zinc/kg for piglets, sows, and all fish species other than salmonids; and 120 mg zinc/kg for other species. Permitted maximum copper contents animal feed in the EU (Regulation 2018/1039) are: 15 mg copper/kg for bovines (cattle) before the start of rumination; 30 mg copper/kg for other bovines (cattle); 15 mg copper/kg for ovines (sheep); 15 mg copper/kg for caprines (goats); 150 mg copper/kg for piglets suckling and weaned up to 4 weeks after weaning; 100 mg copper/kg for piglets from 5th week after weaning up to 8 weeks after weaning; 50 mg copper/kg for crustaceans; and 25 mg copper/kg for other species.
Yu et al. (2017) theorised that certain forms of heavy metals (as stable metal compounds that do not release free metal ions) may provide nutrition to food-producing animals but not be toxic to bacteria, and hence their use in feed would not co-select for resistance in bacteria (Yu et al., 2017). There does not appear to be any evidence supporting this hypothesis.
Other uses of heavy metals include use in livestock footbaths to treat and prevent the spread foot infections such as digital dermatitis (Bell et al., 2014; Yu et al., 2017) and wound dressings (Wales & Davies, 2015). As discussed in more detail later in this report, following concerns over therapeutic use of zinc in animal production potentially leading to an increased prevalence of livestock associated methicillin-resistant Staphylococcus aureus (LA-MRSA) zinc is now only permitted in the EU and UK at concentrations up to 150 ppm for nutritional use (Veterinary Medicines Directorate, 2022). Copper is the principal biocidal component of anti-fouling paints used in aquaculture to reduce the growth of attached organisms on fish cages and nets (Burridge et al., 2010; Guardiola et al., 2012). Copper has also been studied as a possible antimicrobial alternative to stainless steel surfaces in food production and processing (Pontin et al., 2021). The use of silver and zinc nanoparticles as antimicrobial controls for a wide range of applications, including in food animal production, have received considerable attention in recent years (McDonnell & Russell, 1999; Maillard & Hartemann, 2012) and were included in this review. A list of heavy metals considered in this review is shown in Table 2.
Table 2: Heavy metals that were considered or excluded from this review.
| Essential metals (Authorised in animal feed and drugs or * antimicrobial control) |
Non-essential metals (Considered contaminants/ undesirable substances), excluded from review |
|---|---|
| Cobalt (Co) Copper (Cu) Chromium (Cr) Iron (Fe) Manganese (Mn) Molybdenum (Mo) Selenium (Se) Silver (Ag)* Zinc (Zn) |
Arsenic (As) Cadmium (Cd) Mercury (Hg) Lead (Pb) |
Other metals (cadmium, lead, mercury) have no established biological functions and are considered as contaminants/undesirable substances (Hejna et al., 2018). As requested in the FSA specification they were not reviewed in this study. Arsenic has been intentionally used in animal feeds and drugs (to reduce coccidial infection and promote growth) in the past (Silbergeld & Nachman, 2008; Rensing et al., 2018) but it is now banned in most countries, including the UK, the EU 27, and USA, due to concerns on its potential to cause harm to human health and to the environment and was not included in this review.
Mechanisms of action of biocides and heavy metals and similarities in resistance to antimicrobials
Since biocides and their applications are diverse, so are the mechanisms of action of different biocides on bacteria, and consequently on antimicrobial-resistant bacteria and their genes. Biocides have been used extensively in a wide range of applications to control and kill bacteria (Jones & Joshi, 2021). Furthermore, the same biocide can be used for different applications, as the action is concentration dependent and most bacteriostatic and bactericidal actions depend on the concentration used. There are multiple mechanisms of action involving different targets that have been described and they can (i) interfere with the replication of nucleic acids, (ii) interfere with protein synthesis, (iii) alter the structure and function of cell wall, (iv) increase permeability and disrupt the cytoplasmic membrane, and (v) inhibit intermediate metabolic pathways (Liwa & Jaka, 2015).
Quaternary Ammonium Compounds (QAC)s, alcohols, and biguanides lead to bacterial cell lysis with their mode of action involving interactions with bacterial outer membranes and with the cytoplasmic membrane which causes loss of integrity and leakage of the intercellular components and lysis (Jones & Joshi, 2021). Another group of biocides (protonophores) which include the weak acids, such as citric acid and benzoic acid, interfere with the pH balance of cells causing cytoplasm acidification and disruption of the proton-motive force (PMF). This interferes with the metabolic generation of energy and the cell starts to die. Oxidising biocides (such as hydrogen peroxide and ozone) have a rapid killing action by oxidising organic materials by releasing free radicals or by halogenating molecules within the cell. Other electrophilic biocides (such as glutaraldehyde and formaldehyde) cause enzyme inactivation by interaction with cellular components by covalent linkage (Amjad, 2010). The heavy metals, silver and copper, also have this type of mechanism. Mechanisms of antimicrobial action of and resistance/reduced susceptibility to silver have been described by Maillard & Hartemann (2012).
As discussed further in this report, the widespread use of biocide products in diverse applications often used at inappropriate concentrations has been of some concern since such usage may cause development of AMR in some bacteria (Wesgate et al., 2016).
Heavy metals may be toxic to bacteria. As previously mentioned, several possible modes of antimicrobial action of heavy metals have been reported (Lemire et al., 2013). These are: (i) protein dysfunction, (ii) production of reactive oxygen species (ROS) and antioxidant depletion, (iii) impaired membrane function, (iv) interference with nutrient uptake, and (v) genotoxicity. These mechanisms have been reviewed by Lemire et al. (2013).
The following factors influence the efficacy of antimicrobial agents and the resistance/tolerance of bacteria to such agents (whether biocides and/or heavy metal) (SCENIHR, 2009; Wales & Davies, 2015; VKM, 2016):
- Innate or intrinsic resistance of bacteria, such as the presence of, or accessibility to the target of the agent.
- Number and location of bacteria.
- Age of bacterial community.
- State: vegetative cells or spores.
- Concentration and potency of the antimicrobial agent (concentrations below the minimum inhibitory concentration (MIC) may co-select AMR).
- Physical and chemical factors (for example, pH, temperature, salt, mode of application/contact).
- Organic and inorganic materials.
- Duration of exposure (time).
- Attachment of bacteria and presence and state of biofilms.
The influence of these factors on resistance/tolerance will be discussed in further sections of this report.
Many bacteria have evolved mechanisms of resistance to toxic agents. These mechanisms of resistance may provide resistance to other antimicrobials, thereby leading to co-selection for AMR. There are several similarities and differences between antibiotic and biocide/heavy metal resistance/tolerance as shown in Table 3.
Table 3: Similarities and differences between antibiotic and biocide/heavy metal resistance/tolerance (adapted from Weber et al., 2019).
| Similarities | Intrinsic resistance (for example, spores are resistant to alcohols) and extrinsic resistance (for example, efflux pumps for heavy metals) are well described. Acquired mechanisms of resistance are similar (for example, impermeability, efflux pumps). Biofilms impair inactivation/killing. Inactivation is dependent on the concentration and duration of contact with the antibiotic, biocide, or heavy metal. |
|---|---|
| Differences | Most antibiotics inhibit a specific target in a biosynthetic process. Most biocides have multiple concentration-dependent targets, with subtle effects occurring at low concentrations and more damaging ones at higher concentrations. |
Horizontal Gene Transfer (HGT) of resistance genes
Resistance genes (whether ARGs or biocide resistance genes (BRGs) or heavy metal resistance genes [HMRGs]) in bacteria can be transferred to other bacteria through Horizontal Gene Transfer (HGT). Thus, commensal non-pathogenic bacteria with resistance genes can act as a reservoir for ARGs, BRGs, and HMRGs and transfer resistance to non-resistant human pathogenic bacteria (Bengtsson-Palme, 2017). HGT is driven by mobile genetic elements (MGEs), such as plasmids, integrons, transposons, and staphylococcal cassette chromosome elements that facilitate the movement, transfer, and integration of genes between cells (Bennett, 2008). Resistance genes are not always associated with cultivable ‘live’ bacteria (Figure 1). Viable but non-culturable bacteria (VBNC) may express genes after “lethal” treatments (James et al., 2021). Non-cellular ARGs, which covers genes encapsulated in membrane vesicles (MVs), bacteriophages, or gene transfer agents (GTAs), can persist after disinfection, and can transfer to recipient bacteria in the absence of a live donor bacteria (Woegerbauer et al., 2020; James et al., 2021). The frequency of HGT largely depends on the properties of the MGEs, MVs, or bacteriophages, the characteristics of the donor and recipient populations, and the environment (Verraes et al., 2013; Rossi et al., 2014). As will be discussed in further sections of this report biocides and heavy metals may influence HGT.
Figure 1: Forms and origins of resistance genes quantified by molecular biology approaches.
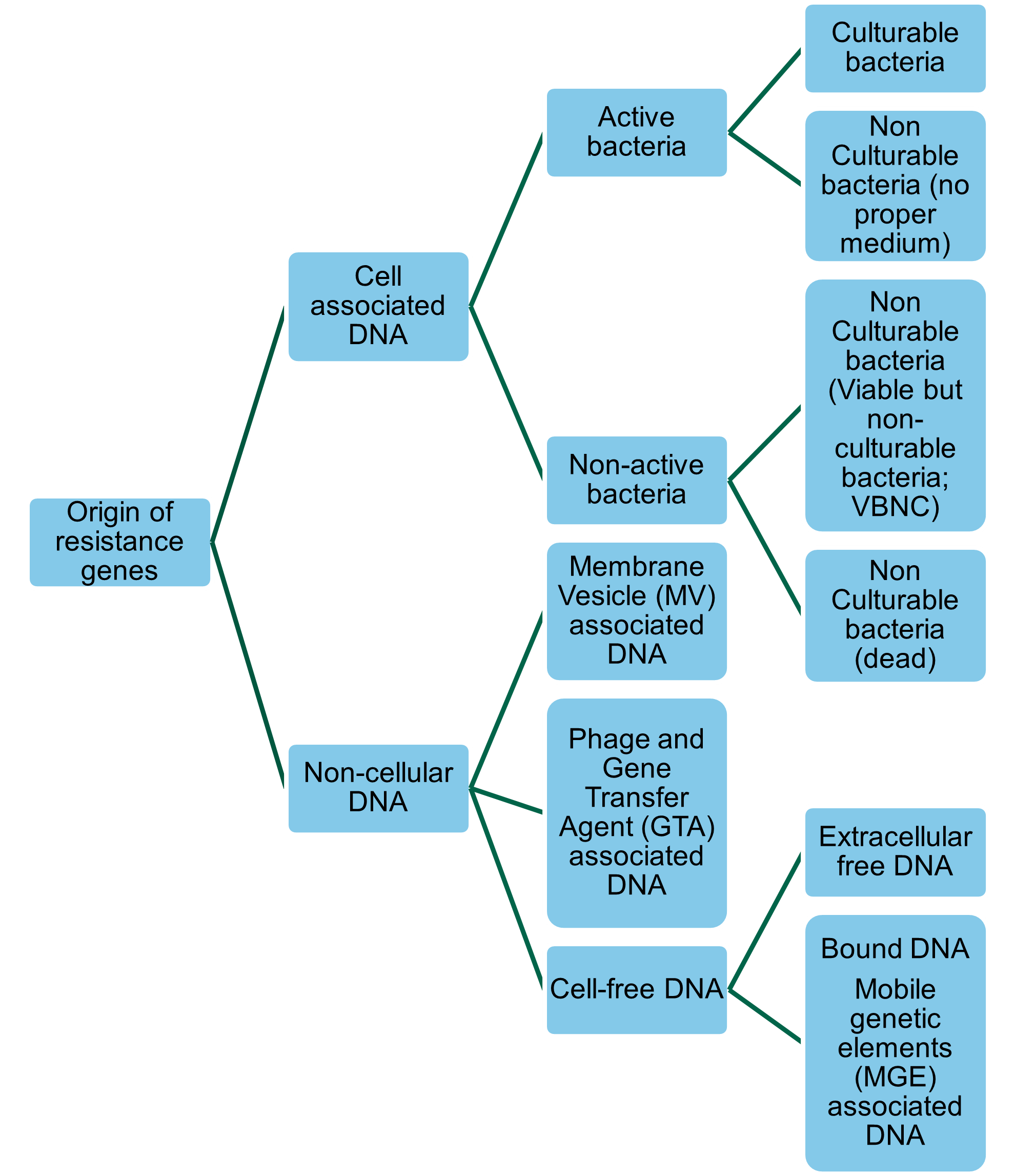
A more detailed discussion of HGT mechanisms can be found in section 13 of this report.
Role of biocides and/or heavy metals in co-selecting AMR
Co-selection mechanisms for biocides and/or heavy metals and clinically- as well as veterinary-relevant antibiotics have been described by Seiler & Berendonk (2012), Wales & Davies (2015), Donaghy et al. (2019), Davies & Wales, (2019), Cheng et al. (2019), and EFSA BIOHAZ Panel (2021), amongst others. There are two main types of related resistance co-selection mechanisms:
- Cross-resistance – where resistance is due to physiological adaptations that provide resistance to a number of toxic agents (such as biocides and antibiotics), examples being efflux pump upregulation, over expression, or reduced cell wall/membrane permeability.
- Co-resistance/co-transfer – where resistance to different toxic agents is dissimilar but there is a genetic link between resistance to different agents, such as the co-location of different resistance genes on the same MGE (mobile genetic elements), such as plasmids but also on chromosomes. Because of the genetic linkage between such resistance, exposure to any of these groups of antimicrobials, or any combination of them, could co-select for the maintenance of the whole MGE and all its associated resistance phenotypes.
Thus, there are some phenomena that confer reduced susceptibility both to antibiotics and to biocides and/or heavy metals (Wales & Davies, 2015; Donaghy et al., 2019; Cheng et al., 2019). These phenomena may be normally present (intrinsic) in the bacteria, or readily acquired by mutation or genetic transfer under appropriate conditions (Wales & Davies, 2015; Donaghy et al., 2019). Phenomena such as spore formation, biofilm formation, nutrient stress responses, low cell wall permeability, and efflux pumps (transport proteins involved in the extrusion of toxic substrates from within cells into the external environment [Webber & Piddock, 2003]) are resistance mechanisms that may enable bacteria to resist antibiotics, biocides, and/or heavy metals (Wales & Davies, 2015; Donaghy et al., 2019). Efflux pumps may expel a broad range of unrelated and structurally diverse compounds including antibiotics, biocides, and/or heavy metals. Thus, whether intrinsic or acquired, bacteria possessing efflux pumps have substantial potential for cross-resistance to antibiotics, biocides, and/or heavy metals, though this does depend on the nature of the efflux pump (Webber & Piddock, 2003; Wales & Davies, 2015).
Resistance may be acquired through the release of resistance genes in MGEs. They may potentially allow some proportion of the bacterial population to survive an otherwise terminal challenge, increasing the risk of selection of organisms permanently adapted to the antimicrobial agent (Wales & Davies, 2015). There can be a genetic link between resistance to different agents (co-resistance) through the co-location of resistance genes on MGEs (Bloomfield, 2002; Wales & Davies, 2015; Ciric et al., 2011).
Resistance in many antimicrobial-resistant bacteria is encoded by genes that are carried on large conjugative plasmids. These plasmids typically contain multiple ARGs as well as genes that confer reduced susceptibility/tolerance to biocides and/or heavy metals (Gulberg et al., 2014). An example of co-resistance are class 1 integrons, which encode a QAC efflux mechanism (qacEΔ1) plus sulphonamide resistance (sul1) and variable other ARGs (Carattoli et al., 2001). The co-existence of blaCTX-M (an ARG encoding resistance to 3rd generation cephalosporins, critically important antimicrobials [CIAs]) and oqzAB (an efflux pump mediating MDR) and pco and sil operons (encoding copper and silver tolerance, respectively) have been reported on the same plasmid isolated from E. coli in food-producing animals (Fang et al., 2016; Zingali et al., 2020). The co-existence of heavy metal tolerance operons (including silA, encoding for tolerance to silver) in plasmids harbouring ARGs including blaCTX-M-2 and the quinolone resistance gene, qnrB isolated from Salmonella spp. from Brazilian poultry has been observed (Ferreira et al., 2019; Galetti et al., 2021). Plasmids isolated from E. fergusonii from poultry have been observed to harbour ARGs and heavy metal tolerance operons (Galetti et al., 2019).
An analysis of the co-occurrence of resistance/tolerance genes to antibiotics, biocides, and metals by Pal et al. (2015) concluded that plasmids provide limited opportunities for biocides and metals to promote HGT of AMR through co-selection (though this was more common in bacteria of animal origin), whereas greater possibilities exist for indirect selection (and therefore clonal selection via chromosomal BRGs and HMRGs).
There is evidence that zinc and/or copper may co-select for LA-MRSA due to co-location of the zinc/copper HMRG czrC and the methicillin resistance gene mecA within the staphylococcal cassette chromosome (SCC) SCCmec element (Aarestrup et al., 2010; Cavaco et al., 2010; Xue et al., 2015; Argudín et al., 2016; Hau et al., 2017; Poole et al., 2017; Jensen et al., 2018). SCCmec is a MGE that carries the mecA gene (or its homologue mecC encoding resistance to methicillin and all β-lactam drugs) and other functional genes (including HMRGs), and can transfer to other Staphylococcus spp.
The persistence of bacteria in food production environments is often associated with their biofilm forming ability. Biofilms are complex structures formed by different or single types of bacteria adhering to surfaces which may enhance resistance to different antimicrobial agents (Uruén et al., 2021). Biofilms have an extracellular matrix that provides a diffusion barrier and an enhanced medium for bacterial signalling and genetic exchange, plus a potential site for neutralisation or binding of chemical agents and an extracellular site for sequestration of metal ions (Wales & Davies, 2015; Donaghy et al., 2019). Once a biofilm forms, bacteria become more resistant to external factors. Bacterial biofilms have been well documented to be highly resistant to antimicrobials, whether biocides or antibiotics (Maillard, 2020). The presence of multiple species may allow for HGT of resistance genes between different bacteria (Allen et al., 2016). Biofilms can generate a state of hypermutability (capability for excessive mutation) in part due to stress and slower growth that stimulates the development of resistance which may co-select for AMR (Yu et al., 2017; Uruén et al., 2021).
There is evidence that some adaptations that enable resistance to antimicrobial agents may result in associated costs to the organism, usually termed “fitness cost”. An example is broad substrate efflux pumps, which consume cell energy resources and indiscriminately remove some useful metabolic substances from the cell (Wales & Davies, 2015; Davies & Wales, 2019). Plasmids encoding resistance to biocides or heavy metals plus antibiotics have been cited as another example (Gulberg et al., 2014). It has been reported that compensatory mutations can arise which offset such plasmid fitness costs (Hall et al., 2021). This has been reported for Pseudomonas fluorescens when acquiring a conjugative plasmid which encodes tolerance to mercury with acquisition resulting in the formation of small colony variants. After repeat passage transconjugants (bacteria that had incorporated DNA from others via conjugation) resumed normal colony size, which was linked to chromosomal mutation (Hall et al, 2019).
A further mechanism that may be relevant to co-selection is the influence of biocides and/or heavy metals on gene transfer (Wales & Davies, 2015; Davies & Wales, 2019). There is some evidence that while some biocides at sub-inhibitory concentrations may inhibit gene transfer, others may increase the efficiency of gene transfer.
Maertens et al. (2019) observed that sub-inhibitory concentrations of a QAC (benzalkonium chloride) had no effect on the conjunctive transfer of ARGs in E. coli originating from poultry. While sub-inhibitory concentrations of cetrimide (alkyltrimethylammonium bromide, a mixture of three QACs) have been observed to increase the transduction of plasmid pWG613 via a bacteriophage in S. aureus (Pearce et al., 1999). Likewise, sub-inhibitory concentrations of chlorhexidine (24.4 μg/L), gentamicin (0.1 mg/L) and sulphamethoxazole (1 mg/L) have been observed to significantly increase the frequencies of transfer of antibiotic resistance in E. coli by conjunction, while other biocides had no effect (Jutkina et al., 2018). Experiments involving field (sewage) bacterial communities of E. coli showed that the efficiency of conjugative transfer between genera may be enhanced in the presence of sub-inhibitory concentrations of biocides, namely free chlorine (0.1−1 mg/L), chloramine (0.1−1 mg/L), and hydrogen peroxide (0.24−3 mg/L) (Zhang et al., 2017a).
These studies suggest that the persistence of low concentrations of some biocides in the environment may accelerate the transfer of ARGs. Persistence will depend on the nature of the biocide. In all cases exposure to biocide concentrations higher than the MIC significantly suppressed transfer of ARGs. Heavy metals, such as copper and zinc, have been reported to facilitate HGT of ARGs in water (Zhang et al., 2018; Cheng et al., 2019; Wang et al., 2020). Whether they do in other environments appears not to have been studied. Our literature search has not identified any studies that have specifically looked at the effect of sub-inhibitory concentrations of biocides on gene transfer under the field conditions present in food animal production. Many studies appear to report an association or correlation in BRGs and/or HMRGs and ARGs due to no clear evidence of co-selection mechanisms.
Davies & Wales (2019) have postulated that, given that there is evidence that low concentrations of antimicrobials, whether antibiotics or biocides, elevate the rate of random mutations in exposed bacterial populations (Cogliani et al., 2011) resulting in spontaneous mutants showing cross-resistance to biocides and antibiotics. According to Maillard (2020), mutations resulting from biocide exposure have mainly been investigated with triclosan, but some studies have looked at other biocides, such as QACs. A laboratory study observed that one single exposure to the working concentration of certain biocides (one a mixture of aldehydes and QACs, one a halogenated tertiary amine compound) may provoke the selection of mutant Salmonella Typhimurium with an efflux mediated multidrug resistance (Whitehead et al., 2011).
Role of concentration of biocides and/or heavy metals in co-selecting AMR
For selection of biocide-resistant bacterial strains to occur, some proportion of the population would be expected to survive the application of biocides. The mode of use of biocides would therefore appear to offer fewer opportunities for survivor selection (Wales & Davies, 2015; Donaghy et al., 2019), compared with heavy metals. Biocides are intended to be lethal/inhibitory, usually after a single application, so are used in the field at concentrations that are higher than the MIC determined in the laboratory. In addition, the Minimum Bactericidal Concentration (MBC) may be determined, which is the lowest concentration of an antimicrobial agent required to kill 99.9% of bacteria over a fixed, somewhat extended period, such as 18 hours or 24 hours, under a specific set of conditions. Whereas the MIC test demonstrates the lowest level of antimicrobial agent that prevents growth, the MBC demonstrates the lowest level of antimicrobial agent resulting in microbial death. The use of biocides in the presence of heavy organic soiling or with diluting water containing interfering organic or mineral substances may produce marked reductions in efficacy even at recommended application concentrations. This may occur on farms or in aquaculture and may reduce effective inhibitory concentrations to sub-inhibitory in practice (Wales & Davies, 2015). For example, the efficacy of the use of biocides where heavy soiling is present, such as their use on vehicle wheels and undercarriages, has been questioned by Maillard (2018). Furthermore, some biocides (such as organic acids and EOs) may be used in practice at sub-inhibitory concentrations (below MICs) in feed and water as growth promoters and for pathogen control (Wales & Davies, 2015).
Low concentrations of antimicrobials in the environment may provide resistant strains of bacteria with a competitive advantage since they may be able to grow in such environments faster than non-resistant strains. The minimum selective concentration (MSC) has been defined as the lowest concentration of an antimicrobial at which resistance is positively selected or co-selected. As highlighted by FAO/WHO, 2019 there are little data on what these threshold values should be in order to inform suitable standards for biocide and metal concentrations in food animal production. There is evidence that the MSC is affected where species of bacteria are embedded within complex communities, such as animal faeces, and may be higher than single strain based estimates (Klümper et al., 2019). FAO/WHO (2019) note that the body of evidence to establish such thresholds is likely to take a considerable time to accumulate. Some studies which have been carried out on MSCs for heavy metals in manure, slurry, and soil are discussed later.
In two reviews Kampf examined published evidence on the cross-resistance of Gram-positive (Kampf, 2018) and Gram-negative (Kampf, 2019) bacterial species to biocides. He concluded that there is evidence that sub-inhibitory concentrations of benzalkonium chloride (a QAC used as a sanitiser) and chlorhexidine (a biguanide used as an antiseptic and disinfectant) may co-select for AMR in both Gram-positive and Gram-negative bacteria. There was evidence for sodium hypochlorite cross-resistance in Gram-negative species, but not Gram-positive. In contrast there is no evidence that cross-resistance to antibiotics has been described after low level exposure to glutaraldehyde, ethanol, propanol, peracetic acid, povidone iodine, and polyhexanide in Gram-positive and Gram-negative bacteria species. Studies such as Molina-González et al. (2014) have reported that sub-inhibitory concentrations of trisodium phosphate, sodium nitrite, and sodium hypochlorite may result in increased AMR in strains of S. enterica. Other studies (Soumet et al., 2012) have observed that sub-inhibitory concentrations of other QACs (didecyl dimethyl ammonium chloride (DDAC) and dioctyl dimethyl ammonium chloride (OCDAC)) may co-select for AMR. While Thomas et al. (2000) observed, in a laboratory study, that repeated exposure to sub-inhibitory concentrations of chlorhexidine created stable resistance in Pseudomonas aeruginosa, there was no cross-resistance to any antibiotics tested, although there was some resistance to benzalkonium chloride.
Alternatively, there is the possibility that intact BRGs could survive the application of biocides on surfaces via MGE and transfer to other bacteria via HGT. There appears to be little literature that considers this possibility. In addition, such exposure could trigger a SOS response in bacteria (a response to DNA damage) and this response has been associated with generation of genetic diversity with AMR variants through the formation of reactive oxygen species and mutagenesis. Whether exposure to some biocides can lead to specific or multiple resistances to antibiotics (including CIAs) used in clinical settings remains to be elucidated. The activation of the SOS response has been found to be associated with the formation of biofilms perhaps due to the slower growth rates involved although antibiotic resistant variants may emerge. A further consideration is that the SOS response is important for prophage activation. Activation of the SOS response could therefore result in the release of phage particles which many carry ARGs and thereby contribute to HGT (Diard et al., 2017). There appears to be little literature that considers any of these possibilities.
Unlike biocides, heavy metals are often used at sub-inhibitory concentrations providing more potential for resistance and co-selection of AMR to emerge (Wales & Davies, 2015).
Aims/objectives of this study
The review question was:
“Do biocides and/or heavy metals used in food animal production have an impact on the development of AMR in the food chain?”
The review was structured and aimed at addressing the following key questions (terms of reference) provided by the FSA:
- Is there evidence in the literature to show that biocides and/or heavy metals used in food animal production have an impact on the development of AMR?
- How long are biocides and/or heavy metals (used in food animal production) able to persist in animal production environments and how does this impact on the development of AMR and associated risks?
- What evidence from the literature is there that biocide and/or heavy metal associated AMR enters the food chain through products of animal origin or as a result of crop contamination?
- Is there a potential risk to the consumer from AMR acquired through the use of biocides and/or heavy metals in food animal production?
A central question was whether the release of biocides (in particular disinfectants) and/or heavy metals from food animal production has the potential to create local concentrations where AMR can emerge and spread (as bacteria or genes) and whether this presents a potential risk to the consumer as a result.
A systematic review approach was taken to the literature search (Figure 2). Because of the paucity of specific published studies on this topic a narrative critical review approach was taken to the review of the publications identified.
The review question was:
“Do biocides and/or heavy metals used in food animal production have an impact on the development of AMR in the food chain?”
The key elements of the question (PIO): Population (P), Intervention (I), and Outcome (O), were:
- The population of interest included pathogenic and non-pathogenic antimicrobial-resistant bacteria and their resistance genes. It excluded microorganisms other than bacteria, such as viruses, fungi, and parasites.
- All biocide or heavy metal interventions used in food animal production.
- Relevant outcome measures for interventions were, what impact did the intervention have on antimicrobial-resistant bacteria and ARGs and resistance.
The review adopted a comprehensive search strategy considering all available evidence in the public domain, including peer-reviewed articles, grey literature (for example, government and industry reports), relevant government reports (for example, FSA published studies, ACMSF reports), European and International literature (for example, the EFSA Scientific Opinions, WHO reports, SCENHIR) up to February 2023. This included previously published systematic and critical reviews, and risk assessments, as well as primary research.
The primary source databases searched were Web of Science, Scopus, and MEDLINE. These bibliographic databases are widely used and include food production, agriculture, public health, and food safety subject areas. The initial searches were restricted to records published from the 1st January 1990 to the 26th July 2022. Further searches were carried out during the course of the study to ensure the review was as ‘up to date’ as possible. The last search being carried out to identify any publications prior to 17th February 2023. The finalised keywords were agreed with the Food Standards Agency and were:
co-selection OR “antimicrobial resistance” OR “antimicrobial resistant” OR “antibiotic resistance” OR “antibiotic resistant” OR “drug resistant” OR “drug resistance” OR “multidrug resistant” OR “multidrug resistance” OR “multi resistance” OR “multi resistant” OR ABR OR AMR OR MDR OR MAR OR AMRG
AND
antiseptic OR biocide* OR disinfectant* OR sanitizer* OR sanitiser* OR “essential oil*” OR “heavy metal*” OR antifouling
AND
“food animal production” OR fish OR seafood OR aquaculture OR salmon OR trout OR cow OR cattle OR dairy OR pig OR swine OR sheep OR lamb OR poultry OR chicken OR turkey OR livestock OR food OR manure OR fertiliser OR feed OR crop* OR “ground water” OR soil OR bedding
Focused Google and Google Scholar searches were used to identify relevant grey literature.
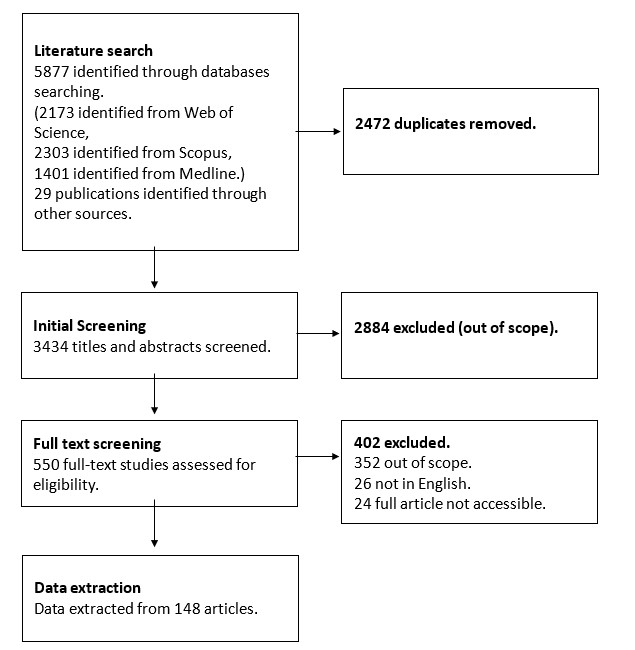
A total of 5,877 citations were initially identified of which 2,173 citations were identified in Web of Science, 2,303 in Scopus and 1,401 in MEDLINE. There was considerable overlap between the databases with 2,472 duplicates leaving 3,405. An additional 29 records were identified through focused Google and Google Scholar searches, searching within relevant publications, and through contact with authors. For all searches, citations and abstracts were uploaded from each of the electronic databases into Covidence (an online tool for systematic reviewing). The following exclusion criteria were applied:
- They contained no relevant data on the impact of biocides and/or heavy metals used in food animal production on the development of AMR.
- Measured irrelevant population (viruses, fungi, and parasites), interventions (biocide not used in food animal production [for example, healthcare]; used for their surfactant properties, antimicrobial peptides [for instance, bacteriocins]; or undesirable heavy metals (such as arsenic [As], cadmium [Cd], mercury [Hg], lead [Pb]), outcomes (did not include impact on antimicrobial-resistant bacteria or genes).
- Were in a language other than English.
The criteria were independently applied to the abstract of each paper by at least two members of the five-member project team. For each citation, a consensus was reached that the citation is relevant for inclusion. Arbitration by a third member of the project team was used to settle conflicting appraisals. A total of 3,434 titles and abstracts were screened, and 2,884 references excluded. Full texts were obtained for all abstracts that passed the inclusion criteria.
A total of 550 publications were considered relevant by title and abstract and full texts collected for second screening. This number was reduced to 148 publications from which some data were extracted, with 402 articles being excluded because they were not in English, the full article was not accessible, or the article was out of scope. An in-depth content analysis of the selected articles was carried out. With the key elements of interest from each paper extracted. To synthesise the data extracted and evaluate its quality a narrative approach was used. This was used to: a) develop a synthesis of findings of the studies, b) investigate relationships within and between studies, and c), evaluate the degree of robustness of the synthesis.
Figure 2: Flow diagram of the selection and exclusion of articles related to the scope of this review.
Existing reviews
As well as the specific studies that have been reviewed, the literature search identified a number of relevant reviews on this subject, or aspects of this subject, notably Wales & Davies (2015), these reviews are listed in Table 4.
Table 4: Key relevant reviews on the impact of biocides and heavy metals on AMR related to food animal production (in chronological order).
| Title of review | Reference |
| Assessment of the antibiotic resistance effects of biocides | SCENIHR (2009) |
| Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture | Seiler & Berendonk (2012) |
| Antibiotic-resistant bacteria: A challenge for the food industry | Capita & Alonso-Calleja (2013) |
| Zinc and copper in animal feed - development of resistance and co-resistance to antimicrobial agents in bacteria of animal origin | Yazdankhah et al. (2014) |
| Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens | Wales & Davies (2015) |
| Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance | Cabello et al. (2016) |
| Supplementation of pigs diet with zinc and copper as alternative to conventional antimicrobials | Dębski (2016) |
| Assistance in the update of the systematic literature review (SLR): “Influence of copper on antibiotic resistance of gut microbiota on pigs (including piglets)” | Van Noten et al. (2016) |
| EMA and EFSA Joint Scientific Opinion on measures to reduce the need to use antimicrobial agents in animal husbandry in the European Union, and the resulting impacts on food safety (RONAFA) * | EMA & EFSA (2017) |
| Antimicrobial resistance and its association with tolerance to heavy metals in agriculture production | Yu et al. (2017) |
| The rising tide of antimicrobial resistance in aquaculture: sources, sinks and solutions | Watts et al. (2017) |
| Methicillin-resistant Staphylococcus aureus: livestock-associated, antimicrobial, and heavy metal resistance (particularly with reference to South Africa) | Dweba et al. (2018) |
| Environmental and public health related risk of veterinary zinc in pig production-using Denmark as an example | Jensen et al. (2018) |
| Resistance to metals used in agricultural production | Rensing et al. (2018) |
| Joint FAO/WHO Expert Meeting in collaboration with OIE on foodborne antimicrobial resistance: Role of the environment, crops and biocides | FAO/WHO (2019) |
| Antimicrobial resistance on farms, including the role of disinfectants in resistance selection | Davies & Wales (2019) |
| Selection and dissemination of antimicrobial resistance in Agri-food production | Cheng et al. (2019) |
| The role of emerging organic contaminants in the development of antimicrobial resistance | Alderton et al. (2021) |
| Scientific opinion on the role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain ** | EFSA BIOHAZ Panel (2021) |
| Factors influencing the transfer and abundance of antibiotic resistance genes in livestock environments in China | Wang et al. (2022b) |
| Interactions and associated resistance development mechanisms between microplastics, antibiotics and heavy metals in the aquaculture environment |
Li et al. (2022b) |
| Evaluating the impact of heavy metals on antimicrobial resistance in the primary food production environment: A scoping review | Anedda et al. (2023) |
|
* This EMA and EFSA opinion did not specifically consider biocides or heavy metals but did discuss their role as alternative antimicrobials and risks for co-selection. |
The majority of these reviews are focussed on land-based food animal production. These reviews have repeatedly highlighted the lack of clear in-field evidence on the role of non-antibiotic drivers in co-selecting AMR in the environment. These reviews (for example, Davies & Wales, 2019) highlight that there is scarce evidence on the efficacy or selective effect of currently used farm and hatchery biocides on the survival or emergence of organisms with increased biocide tolerance or AMR. Investigations of bacterial strains recovered from the field appear to show some evidence of associations/correlations between biocide use and increased AMR although clear evidence for causal links is relatively scant. These studies were limited in terms of the organisms examined - most choose a specific bacterial spp., whether zoonotic or not, and ignored the rest of the microbial community.
The SCENIHR (2009) classified biocides according to their intrinsic potential for generating resistance/tolerance. Some biocides, due to the nature of their interactions with the bacteria, would be more prone to induce resistance/tolerance. SCENIHR considered that this group of high-risk biocides contains the QAC, biguanides (i.e., surface active agents), phenolics, and metallic salts. On the other hand, highly reactive biocides (for example, oxidising and alkylating agents) present a low risk of emergence of bacterial resistance, and that reduced susceptibility to these biocides results mainly from their inappropriate use. Several other biocides (isothiazolones, anilides, diamidines, inorganic acids and their esters or alcohols) have been classified as medium-risk in terms of emergence of bacterial resistance.
Few of these reviews have considered the impact of the use of biocides and/or heavy metals on AMR in aquaculture. Reviews by Seiler & Berendonk (2012), Cabello et al. (2016), Watts et al. (2017), EFSA BIOHAZ Panel (2021), and Li et al. (2022b) in relation to heavy metals mention heavy metals (and biocides, in the case of Cabello et al., 2016) as potential drivers for co-selection of AMR but they do not cite any specific studies that have addressed the use of heavy metals in feed or other compounds (biocides) used in aquaculture on co-selection of AMR. Nor has our literature review identified any compelling studies that have mapped the use of heavy metals (whether in feeds or in other uses) or biocides in aquaculture with co-selection of AMR.
Impact of biocides on AMR in food animal production
As highlighted by the SCENIHR (2009), Wales & Davies (2015), Cheng et al. (2019), Donaghy et al. (2019), and Giacometti et al. (2021) amongst others, and confirmed in our literature search, while there is much laboratory experimental evidence on the impact of biocides in selecting antibiotic resistance there are considerably less field data in relation to the food animal production context. As noted in a number of reviews, the efficacy of biocidal action in the field and ability to select AMR may be significantly reduced due to the presence of heavy organic soiling or dilution effects. In general, studies do not appear to have specifically quantified these effects on MICs in the context of AMR co-selection. A few studies (as discussed below) have observed that sustained exposure of livestock-associated bacteria to sub-inhibitory concentration of biocides may result in increased levels of AMR among these bacteria.
There has been a concern regarding the potential for poultry biocides used as antimicrobial processing aids during poultry processing operations to increase AMR (Rhoma et al., 2021). Biocides are used to treat poultry during processing in many countries, though not generally in Europe. Reviewing the evidence on peroxyacetic acid, chlorine, chlorite, and trisodium phosphate as a poultry carcass and meat decontaminant, the European Food Standards Authority Panel on Biological Hazards (EFSA BIOHAZ Panel) concluded that there was no evidence that their use would lead to acquired reduced susceptibility among contaminating bacteria, nor to acquired resistance to antibiotics (EFSA BIOHAZ Panel, 2008).
An investigative laboratory study that repeatedly exposed four strains of L. monocytogenes and S. Typhimurium from poultry to increasing sub-inhibitory concentrations of five decontaminants (trisodium phosphate (TSP), chlorine dioxide (CD), acidified sodium chlorite (ASC), citric acid (CA), and peroxyacetic acid (PA)) demonstrated an increase in resistance in the strains to various antibiotics of the 15 they were tested against (Alonso-Hernando et al., 2009). There was no strong pattern of association, although development of resistance to the aminoglycosides, streptomycin and neomycin was common. Strains exposed to ASC showed a higher number of increased resistances, 13 resistances to the 15 antibiotics against which they were tested than when exposed to TSP (7 resistances), CA (3 resistances), CD (5 resistances) or PA (7 resistances). A laboratory-based investigative study that dipped chicken legs in solutions of TSP, ASC, ascorbic acid (AA), CA, or water (control) showed that E. coli isolated from the samples had a higher level of resistance to ampicillin-sulbactam (treated with TSP), amoxicillin-clavulanic acid (treated with ASC, AA and CA), cefotaxime (treated with TSP), trimethoprim-sulphamethoxazole (treated with AA or CA), tetracycline (treated with CA), ciprofloxacin (treated with ASC, AA, or CA) and nitrofurantoin (treated with TSP) in comparison with controls (Capita et al., 2013).
In their review of evidence on this subject Rhouma et al. (2021) concluded that there is at present some evidence that sub-inhibitory concentrations of biocides may lead to selection, but that at present the published evidence is scare and derived from laboratory-based experiments. They suggested that due to the risk of co-selection that the poultry industry should consider non-chemical physical interventions, such as hot water and steam treatments. We would echo that and note that some poultry processors in the UK are currently employing such physical interventions.
Studies by Randall et al. (2005, 2007) have observed that the use of certain types of biocides commonly used in UK farms can increase bacterial resistance/tolerance to both biocides and to antibiotics. A laboratory-based study (Randall et al., 2005) showed no co-selection effect with ciprofloxacin-resistant strains of E. coli to three commercial disinfectants (a tar oil phenol, which was a blend of high boiling point tar acids and organic acid, an oxidising compound, and a combination of formaldehyde, glutaraldehyde and QAC). There was a slight increase in cyclohexane tolerance among a minority of disinfectant-passaged strains (particularly those subjected to the phenolic biocide). A further laboratory based study (Randall et al., 2007) exposed eight S. Typhimurium isolates (including field isolates and laboratory mutants) to different farm biocides (a tar oil phenol; an oxidising compound; an aldehyde-based disinfectant; or QAC’s). Results differed depending on the biocide and the Salmonella spp. strain tested. Exposure to an aldehyde-based disinfectant reduced susceptibility to the fluoroquinolone ciprofloxacin in some strains. An analysis of proteomes (the complete set of proteins made by an organism) revealed significantly increased expression of the AcrAB–TolC efflux system (responsible for resistance to antimicrobials) after exposure to a tar oil phenol disinfectant. The results showed that single exposure to biocides was insufficient to select for AMR strains. The authors concluded that biocides could be a selective pressure for the selection and/or maintenance of ciprofloxacin-resistant strains in the farm environment in the absence of ciprofloxacin itself.
Nhuyen et al. (2015) observed that the sustained exposure of E. coli and non-typhoidal Salmonella (isolated from farmed animals) to sub-inhibitory concentrations of a commonly used commercial disinfectant containing a mix of benzalkonium chloride (a QAC) and glutaraldehyde used on pork and poultry farms in Vietnam appeared to co-select AMR. Increases in MIC for the biocide were strongly correlated with reduced susceptibility shown by increases in MIC (or decreases in inhibition zone) for ampicillin, tetracycline, ciprofloxacin, and chloramphenicol, and to a lesser extent for gentamicin, trimethoprim/sulphamethoxazole. To investigate whether generic efflux pump expression was responsible for the observed changes, the study treated strains with a generic efflux pump inhibitor and measured the changes in AMR before and after treatment. Results suggested that mechanisms other than efflux pumps were responsible for co-selection.
Davies & Wales (2019) cited unpublished data from the Animal and Plant Health Agency (APHA) that there were concerns that the use of sub-inhibitory concentrations of QACs, as a consequence of cost and staff safety issues, was becoming a common practice in UK poultry hatcheries. They reported that there was evidence that certain quinolone-resistant, hatchery-resident Salmonella spp. strains appeared to have emerged from such situations and subsequently spread to broilers. This evidence does not appear to have been published elsewhere, or any similar studies undertaken.
Many essential oils (EOs), plant compounds, and extracts have been shown to act as antimicrobial agents and promoted as ‘natural’ alternative feed additives to antibiotics (De Souza, 2016; Stevanović et al., 2018; Álvarez-Martínez et al., 2021; Mariotti et al., 2022). While there is much evidence on the efficacy of EOs there are little data on their modes of action (Álvarez-Martínez et al., 2021) and potential to drive co-selection of resistance (De Souza, 2016). De Souza (2016) further concluded in their review on the effects of sub-inhibitory doses of EOs on AMR that EOs were not likely to impose a major hazard. There is some evidence that controlled exposure of bacteria to sub-inhibitory concentrations of EOs can alter and select for AMR (Al-Mnaser & Woodward, 2020), but field studies are lacking. There is evidence that different EOs may have different effects on resistance. A comparison of effects of low concentrations of cinnamon and oregano on resistance in Gram-negative bacilli observed that repeated use of cinnamon had no effect on AMR, but oregano could increase Proteus mirabilis resistance to ampicillin or decrease Serratia marcescens resistance to tetracycline (Becerril et al., 2012). Since EOs are composed of many chemical constituents, it is not surprising that different oils show synergistic or antagonistic effects. EOs have been advocated as alternatives to antibiotics in animal feeds, but studies do not appear to have considered whether their use may be co-selective. Thymus maroccanus (a species of thyme) EO has been shown in vivo to select for AMR in E. coli strains (Fadli et al., 2014). Resistance was associated with an overexpression of an efflux pump related to AcrAB-TolC (a RND-based tripartite efflux pump) in some variants. Sub-inhibitory concentrations of tea tree oil (Melaleuca alternifolia) have been associated with reduced susceptibility to antibiotics in E. coli, S. aureus, MRSA, and Salmonella spp. (McMahon et al., 2007b).
Menthol (an EO) has been suggested as an antibiotic alternative in cattle. A US study (Aperce et al., 2016), on feedlot cattle that had been fed menthol, reported no increased resistance in E. coli isolates to many antibiotics (amoxicillin, ampicillin, azithromycin, cefoxitin, ceftiofur, ceftriaxone, chloramphenicol, ciprofloxacin, gentamicin, kanamycin, nalidixic acid, streptomycin, sulfisoxazole, and sulphamethoxazole). They did observe an increased prevalence of tetracycline-resistant E. coli after 30 days of menthol supplementation (0.3 %) in feed. Similarly, another study by this research team (Murray et al., 2021), reported a trend (though not statistically significant) in increasing resistance with menthol (0.3 %) supplementation (also in combination with zinc [300 ppm]), particularly reduced sensitivity to tetracycline. No mechanistic explanations were pursued in these studies. It is not clear from these studies why a concentration of 0.3 % was used, and trials were not carried out to determine the effect of different concentrations of menthol in feed on co-selection. These studies note that menthol in feed has been shown to promote weight gain in poultry, and there are published studies on its use in fish feed. No other studies appear to have been undertaken to establish whether menthol in feed may select for AMR in other animal species.
It is usually assumed that conventional cleaning and disinfection procedures using biocides that are effective in eliminating non-antimicrobial-resistant bacteria will be equally effective against antimicrobial-resistant bacteria on farms (Davies & Wales, 2019). Davies & Wales (2019) cite some unpublished evidence that LA-MRSA may be more resistant than Salmonella spp., as well as evidence published by Kotb & Sayed (2015). A recent study by Montagnim et al. (2022) highlighted that MDR strains of E. coli may be more resistant to some commonly used farm disinfects than non-MDR strains. The efficacy of cleaning and disinfection regimes in reducing AMR on farms is discussed in reviews such as Davies & Wales (2019).
A strong association was found between frequent disinfection of pens and colonisation of nursery piglets with LA-MRSA in Canada. All LA-MRSA isolates carried at least one QAC BRG, with the most common genotype being qacG qacH smr (Slifierz et al., 2015b). Nursery herds testing positive for LA-MRSA reported more frequent use of disinfectants (as well as zinc therapy), as well as having a higher stocking density. This is one of the few animal food production in-field studies identified in our literature search that have addressed the impact of biocides on AMR. The study did not map the use of specific disinfectants to resistance.
Wieland et al. (2017) observed no association of increased didecyl dimethyl ammonium chloride (DDAC, a QAC) MICs with ESBL/AmpC isolates from poultry but they did observe significant positive correlations for MIC values of DDAC (≤0.36–3.6 mg DDAC l-1) and four other antibiotics (chloramphenicol, florfenicol, piperacillin, sulphamethoxazole + trimethoprim) in E. coli, as well as for 13 antibiotics in enterococci, suggesting that residual QACs may select AMR enterococci.
A similar study showed an association in reduced susceptibility to sodium hypochlorite and AMR in Salmonella spp. from poultry in China (Xiao et al., 2022). Positive correlations between chlorine tolerance and clinical antibiotic resistance to ceftiofur, tetracycline, ciprofloxacin and florfenicol were observed. The most frequently detected chlorine BRG was qacE∆1; thus cross-resistance was likely due to efflux pump over-expression. This gene is also associated with MGEs (such as class I integrons) and co-selection (Hu et al., 2018).
A study of E. coli from pigs, pig carcasses and pork in Thailand (Puangseree et al., 2021) showed a weak correlation between reduced susceptibility to biocides and AMR. Some cross-resistance between benzalkonium chloride (a QAC) and chloramphenicol, ciprofloxacin, sulphamethoxazole, and tetracycline; and chlorhexidine and ciprofloxacin, gentamicin, and streptomycin, was observed.
A study by Cufaoglu et al. (2022) investigated AMR and biocide susceptibility (as well as heavy metals, as will detailed in the next section) amongst E. coli isolates from chicken, cattle, and sheep in Turkey. A high prevalence of AMR were reported amongst isolates, along with a high prevalence of reduced susceptibility to N-alkyl-dimethyl-benzyl-ammonium [a QAC] (26%). In order to determine susceptibility to biocides (and heavy metals), field isolates were deemed tolerant, if the MIC of field isolates was above that of a control E. coli strain, ATCC 25922. This highlights an issue in the literature around breakpoints and how resistance is determined, in this study resistance/reduced susceptibility was reported if the MIC was one doubling dilution above the control and no replication indicated. The authors did report the presence of BRGs in a smaller proportion of isolates tested by PCR assay, with one MDR isolate subject to whole genome sequencing (WGS). Such genes were found in this isolate and were chromosomally located. Whilst HMRGs, BRGs and ARGs were found in the same isolates, there were limited data presented to suggest that the use of biocides specifically were driving AMR.
Other studies on biocides (as discussed below) have not observed any evidence of cross-resistance or co-selection between antibiotics and biocides.
A Brazilian comparison of biocide (sodium hypochlorite and benzalkonium chloride [a QAC]) use on AMR of S. Heidelberg isolated from poultry flocks in 2006 with those isolated in 2016 showed no increase in resistance/tolerance over this time period (to either biocides or antibiotics [with the exception of tetracycline resistance which showed an increase]), suggesting no signs co-selection from biocide use (Bassani et al., 2021).
A survey of field strains of S. enterica from pigs (132 strains) and poultry (125 strains) in Thailand commonly exhibited the multiple antibiotic resistance (MAR) phenotype (42%), but this was not associated with reduced susceptibility to benzalkonium chloride (a QAC) or chlorhexidine among the 257 strains (Chuanchuen et al., 2008).
A study of Salmonella isolates from two commercial US turkey processing plants found that all Salmonella isolates were chlorhexidine tolerant, but no cross-resistance between chlorhexidine and five antibiotics (gentamicin, kanamycin, sulphamethoxazole, streptomycin, and tetracycline) was found in the 130 Salmonella spp. serovars compared in the laboratory (Beier et al., 2011). A series of later studies (Beier et al., 2013; Beier et al., 2019 and Beier et al., 2021) by the same US research group compared biocide tolerance and AMR in strains of E. coli O157:H7, Campylobacter coli, and C. jejuni isolated from cattle, pigs, and poultry, respectively. In all cases no correlation between biocide tolerance and AMR was observed. The beef study (Beier et al., 2013) compared biocide tolerance and AMR of 244 strains of E. coli O157:H7 isolated from cattle carcasses, faeces, and hide. It failed to find any correlation between biocide tolerance and AMR. Although only a low prevalence of AMR isolates was observed (14%), mainly to chloramphenicol, streptomycin, and tetracycline. Fifteen different biocides were tested, including QAC based sanitisers, and organic acids. Tolerance to certain biocides (chlorhexidine or benzalkonium chloride) and antibiotics was observed in some strains. The same group compared the tolerance/resistance of 111 C. coli isolated from pigs and pork meat to 22 biocides and 9 antibiotics, finding that 84 to 96% of strains were resistant to chlorhexidine, and 75% were resistant to tetracycline (Beier et al., 2019). In the poultry study (Beier et al., 2021) the resistance of 96 strains of C. jejuni isolated from the litter of chicken houses to 9 antibiotics and 22 biocides (mainly a range of QAC based sanitisers, but also included formaldehyde) was studied. This study found 13.5%, 12.5%, and 22% of the strains demonstrated reduced susceptibility to ciprofloxacin, nalidixic acid, and tetracycline, respectively. In total, 32% of strains were resistant to chlorhexidine, but not to other biocides; and no cross-resistance was observed between the antibiotics and the 22 biocides (disinfectants). An unrelated laboratory study observed no evidence that tolerance to biocides (benzalkonium chloride (a QAC), chlorhexidine, cetylpyridinium chloride (CPC, a QAC), trisodium phosphate, and sodium dodecyl sulphate) was connected to AMR (resistance to erythromycin and/or ciprofloxacin) in 27 C. coli and 15 C. jejuni isolates from food, animal, human and environmental water sources (Mavri et al., 2012).
A study of four French poultry slaughterhouses observed no increased resistance or cross-resistance in Campylobacter isolates after cleaning and disinfection (Peyrat et al., 2008). Similarly, a study of over 500 isolates from Danish pig slaughterhouses showed there was little evidence of an association between biocide use and AMR in Salmonella spp. (Gantzhorn et al., 2014). There was no increased tolerance in the isolates to the biocides tested (a QAC and an acid/hydrogen peroxide mix) in comparison to a reference strain of S. Typhimurium and there was no evidence that cleaning and disinfection co-selected for antimicrobial-resistant bacteria. Likewise, a study by Bridier et al. (2019) of cleaning and disinfection procedures over three months in a French pig slaughterhouse observed no evidence of reduced susceptibility to biocides of Salmonella spp. isolates or co-selection of AMR in such isolates.
A Belgium survey of disinfectant use and resistance in E. coli in both poultry and pig production observed no indications for the co-selection of AMR through the use of commonly-used biocides (i.e., glutaraldehyde, benzalkonium chloride, formaldehyde, and a formulation of peracetic acid and hydrogen peroxide) in these environments (Maertens et al., 2019). In a further study by this research team (Maertens et al., 2020) the susceptibility of field E. coli isolates from a broiler and pig pilot farm to 14 antibiotics and the 4 disinfectants was monitored over a one-year period. No change in biocide tolerance to these disinfectants was observed and no association was found between biocide use and AMR.
As well as their use for cleaning and disinfecting biocides are used in footbaths and to clean udders of animals used for milk production (SCENIHR, 2009; Wales & Davies, 2015; Donaghy et al., 2019; VKM, 2016). Few studies on the impact of such practices on co-selection for AMR were found in our literature search.
A number of studies have identified a concern that inappropriate application of teat dipping biocides applied to dairy cattle could co-select for AMR. Few studies have demonstrated whether this may occur. A study by Abd El-Aziz et al. (2021) of Streptococcus uberis from bovine clinical mastitis in dairy farms with diverse hygienic interventions in Egypt showed that qac resistance genes were positively correlated with ARGs/AMR phenotypes in the isolates studied. No details were provided on the type of antiseptic used, and no clear evidence of a link between disinfectant use and AMR was demonstrated. A German study (El Behiry et al., 2012) observed no cross-resistance in S. aureus from cows with subclinical mastitis that showed reduced susceptibility to commercial teat dips (nonoxinol-9 iodine complex and chlorhexidine). This suggested that reduced susceptibility to these biocides may be distinct from AMR. An Italian study observed that coagulase-negative staphylococci from milk had a low prevalence of qac genes encoding for disinfectant efflux pumps and there was no evidence of cross-selection (Turchia et al., 2020).
A study by Roedel et al. (2021) on E. coli from broiler meat farms did not find a link between phenotypic biocide tolerance to commonly-used biocides and AMR on those farms. The study determined both the MIC and MBC and determined reduced susceptibility based on the MIC95 or MBC95 (the antibiotic concentration that would inhibit the growth of 95% of the tested bacterial isolates) therefore, arguably providing more robust results. When WGS was undertaken for a small number of isolates’ ARGs (aadA1 and sul1) were found closely linked with qacE∆1 on a class 1 integron and which included a mercury-resistance gene operon. There was an association between blaCMY-2 and sugE(p) (conferring resistance to QACs) found on the same contig (a consensus region of DNA from overlapping DNA sequences from WGS mapping a region of DNA) and which was associated with genes suggestive of localisation on a plasmid.
Biocides and heavy metals (copper and zinc, as discussed in the relevant section of this report) are routinely used in antimicrobial footbaths in commercial dairy farming to prevent lameness caused by bacterial infections (Bell et al., 2014; Yu et al., 2017). A number of different biocides may be used, with Bell et al. (2014) reporting formalin to be commonly used in the UK. Few studies have investigated their impact on AMR co-selection. A study of disinfecting footbaths used in six Norwegian dairy farms found that Serratia marcescens may survive and multiply in these baths but there were no indications of cross-resistance between biocides and AMR in surviving isolates (Langsrud et al., 2003). The contents of these footbaths are usually disposed into slurry tanks, which is likely to lead to soil contamination and a potential driver for co-selection of AMR (Williams et al., 2019).
The effects of biocides and their impact on AMR in the aquacultural environment (whether marine or fresh) appears to be poorly studied. Our literature search identified very few published studies. According to Burridge et al. (2010) compounds used are water soluble and of low toxicity depending on the quantities used. They report that Virkon ®, iodine + detergents, chloramine-T, hypochlorite chlorine dioxide, benzalkonium chloride, Superquats ®, glutaraldehyde, formalin 40%, calcium oxide, calcium hydroxide, sodium carbonate, Creolina, synthetic phenols, halophenols and ethanol are used in Chile in salmon aquaculture. We have not found data on which compounds are used in the UK. Defra currently list the products which have demonstrated effectiveness again aquatic disease through testing in the UK done via The Centre for Environment, Fisheries and Aquatic Science (Cefas). These include many of the products above and interestingly state that testing is done at 4+1 degrees celsius so under industry appropriate conditions.
A study by Romero et al. (2017) has been widely cited in the literature as providing evidence of the co-selection of biocides (and heavy metals) on AMR in seafoods. This study observed multiple /tolerances resistances (to biocides, heavy metals, and antibiotics) in 76% of isolates from a wide range of seafoods. ARGs detected included sul1 (43.33% of tested isolates), sul2 (6.66%), blaTEM (16.66%), blaCTX−M (16.66%), blaPSE (10.00%), blaIMP (3.33%), blaNDM−1 (3.33%), floR (16.66%), aadA1 (20.0%), and aac(6′)-Ib (16.66%) and is of concern given that blaIMP and blaNDM−1 encode resistance to carbapenems (CIAs). The only BRG detected was qacE∆1 (10.00%). These results suggest that exposure to biocides may co-select for AMR. The fish and seafood sampled was purchased at supermarkets and fish markets in the region of Jaen, Spain. While many samples were sea-caught fish; sea bass, salmon, and prawn samples were farmed and showed patterns of tolerance/resistance to biocides and antibiotics. No direct comparison with any pattern of use of biocides during the husbandry of these seafoods was made.
The effects of biocides and their impact on AMR in the aquacultural environment (whether marine or fresh) appears to be poorly studied. Our literature search identified very few published studies. According to Burridge et al. (2010) compounds used are water soluble and of low toxicity depending on the quantities used. They report that Virkon ®, iodine + detergents, chloramine-T, hypochlorite chlorine dioxide, benzalkonium chloride, Superquats ®, glutaraldehyde, formalin 40%, calcium oxide, calcium hydroxide, sodium carbonate, Creolina, synthetic phenols, halophenols and ethanol are used in Chile in salmon aquaculture. We have not found data on which compounds are used in the UK. Defra currently list the products which have demonstrated effectiveness again aquatic disease through testing in the UK done via The Centre for Environment, Fisheries and Aquatic Science (Cefas). These include many of the products above and interestingly state that testing is done at 4+1°C, so under industry appropriate conditions.
A study by Romero et al. (2017) has been widely cited in the literature as providing evidence of the co-selection of biocides (and heavy metals) on AMR in seafoods. This study observed multiple /tolerances resistances (to biocides, heavy metals, and antibiotics) in 76% of isolates from a wide range of seafoods. ARGs detected included sul1 (43.33% of tested isolates), sul2 (6.66%), blaTEM (16.66%), blaCTX−M (16.66%), blaPSE (10.00%), blaIMP (3.33%), blaNDM−1 (3.33%), floR (16.66%), aadA1 (20.0%), and aac(6′)-Ib (16.66%) and is of concern given that blaIMP and blaNDM−1 encode resistance to carbapenems (CIAs). The only BRG detected was qacE∆1 (10.00%). These results suggest that exposure to biocides may co-select for AMR. The fish and seafood sampled was purchased at supermarkets and fish markets in the region of Jaen, Spain. While many samples were sea-caught fish; sea bass, salmon, and prawn samples were farmed and showed patterns of tolerance/resistance to biocides and antibiotics. No direct comparison with any pattern of use of biocides during the husbandry of these seafoods was made.
Overall, the literature on investigations of bacterial isolates recovered from the field appear to show some evidence of associations/correlations between biocide use and increased resistance to antibiotics. Particularly there is some evidence that QACs, such as benzalkonium chloride, that are widely used in food animal production for disinfection of farm environments and equipment, and chlorhexidine, a biguanide used as an antiseptic and disinfectant for example as a dairy teat disinfectant, may co-select AMR, although there appears to be little clear evidence in the literature for causal links in the field. These biocides have also been identified as risks in other reviews, such as SCENIHR (2009). There is clearly a need to establish whether current cleaning and disinfection regimes in use in food animal production in the UK represent a real hazard with respect to the selection of AMR.
Impact of heavy metals on AMR in food animal production
As highlighted by Wales & Davies (2015), Cheng et al. (2019), Donaghy et al. (2019), and Giacometti et al. (2021), while there is some laboratory experimental evidence on the impact of heavy metals on the selection or development/dissemination of AMR, there are considerably fewer field data (though considerably more than on the impact of biocides). In common with the evidence on the impact of biocides, while there are some data showing an association/correlation in resistance, there is little clear evidence for causal links. A summary of in-field studies that have addressed the impact of heavy metals on AMR is shown in Table 5. Evidence is mainly on the supplementation of pig feed with zinc or copper, or the therapeutic use of zinc oxide in pig production. There are little data on the impact of heavy metals on AMR in other forms of food animal production, particularly aquaculture. These studies do not directly address whether an increase in AMR or ARGs as an impact of heavy metal use may in turn lead to an increased risk to public health, but only to the dissemination of AMR and ARGs in the environment.
There is concern that therapeutic use of zinc oxide in pig production (which is used to prevent postweaning diarrhoea) and in feed (as a growth promotor) at high concentrations (as high as 3,000 ppm rather than 50 to 150 ppm required for nutritional use) may co-select for LA-MRSA. This is due to the co-location of the zinc/copper resistance gene (czrC) and methicillin resistance gene (mecA) within the staphylococcal cassette chromosome mec (SCCmec) element (Aarestrup et al., 2010; Cavaco et al., 2010; Argudín et al., 2016; Hau et al., 2017; Poole et al., 2017; Jensen et al., 2018). SCCmec is a MGE that carries the mecA gene [or its homologue mecC] (encoding resistance to methicillin and all β-lactam drugs) and other functional genes, and can transfer to other Staphylococcus spp.
An association between reduced zinc susceptibility and the development of LA-MRSA CC398 in Danish pigs was shown to be a consequence of the frequent presence of czrC in SCCmec (type V) in both pig and human isolates (Aarestrup et al., 2010; Cavaco et al., 2010). Alen et al. (2018) reported an increase in the percentage of zinc tolerant LA-MRSA CC298 isolated from patients of a German university hospital located in a pig farming-dense area between 2000 and 2014 which they associated with the use of zinc in pig feed. Prior to 2009, about half of the LA-MRSA CC398 isolates were zinc tolerant, whereas by 2014 all tested LA-MRSA CC298 isolates were found to be zinc tolerant. Zinc tolerance was found to correlate with the presence of the czrC gene in all cases. A small-scale study from USA confirmed a strong association of S. aureus CC398 from pigs and czrC-positive (Hau et al., 2017). The same study suggests that for certain other lineages (pig-associated LA-MRSA ST5) the contribution of zinc to the emergence of LA-MRSA may be negligible. This may be due to the variations in SCCmec cassettes among LA-MRSA lineages and not all types will harbour SCCmec type V, with ST5 isolates found to carry either SCCmec type III or IV, or untypeable cassettes (Hau et al., 2017). Argudín et al. (2016) demonstrated that the czrC gene was almost exclusively found (98%) in the presence of SCCmec V in both CC398 and non-CC398 LA-MRSA isolates (CC1 and CC97 LA-MRSA). No evidence of zinc contributing to the prevalence of CC398 and CC5 LA-MRSA strains in pig farms in Korea was observed by Eom et al. (2019).
Other genes potentially conferring metal tolerance, including copB (copper), have been found to be present in LA-MRSA and associated with SCCmec and integrons (Argudín et al., 2016). The plasmid pAFS11 obtained from CC398 isolates has been shown to harbour 5 different ARGs and 2 HMRG operons [including copA, encoding copper tolerance] (Feßler et al., 2017). LA-MRSA strains have been described harbouring plasmids carrying tolerance genes for copper (copA and mco HMRGs) and for multiple antibiotics including macrolides, lincosamides, streptogramin B, tetracyclines, aminoglycosides, and trimethoprim (erm(T), tet(L), aadD, and dfrK ARGs) (Gomez-Sanz et al., 2013).
Other studies have shown LA-MRSA in weaner pigs are influenced by exposure to therapeutic doses of in-feed zinc (≥2,000 ppm) when compared to the recommended dietary concentration (100 ppm). Slifierz et al. (2015a) reported was a significant association between the prevalence of LA-MRSA-positive pigs (followed from birth to weaning) and zinc concentration (3,000 vs 100 ppm) at four and five weeks of age. In both groups LA-MRSA-positive animals were similarly infrequent by seven weeks of age in the randomised controlled trial. A further report by the same group found a strong association between the concentration of zinc in the nursery ration and colonisation of nursery piglets with LA-MRSA (Slifierz et al., 2015b). Samples from 390 pigs from 26 farms were compared. Nursery herds testing positive for LA-MRSA reported more frequent use of zinc therapy (≥2,000 ppm in-feed), as well as having a higher stocking density. In this study czrC (a HMRG encoding resistance to zinc and copper) was detected in about two-thirds of isolates in association with a lower susceptibility to zinc compared with czrC-negative isolates.
An in vitro study by Peng et al (2020), investigated the growth of two ESBL-producing E. coli strains carrying blaCTX-M-1, with the gene either on a plasmid or chromosomally- encoded, in pig faecal material containing an increasing concentration of zinc (0-8 mM). Interestingly, expression of the gene increased with increasing zinc concentration. The authors suggest that zinc may be inducing the promoter activity of an insertion element ISEcp1 normally found upstream of the blaCTX-M-1 gene and increased zinc efflux and thus higher level zinc tolerance. Furthermore, at higher zinc concentrations there was a higher proportion of CTX-M-1 resistant E. coli relative to the total flora, but only for the strain where the gene was plasmid encoded. These results suggest that exposure to therapeutic zinc concentrations may give a selective growth advantage to bacteria carrying such plasmid-encoded genes and thereby induce their expression. Such selection does therefore not have to be linked to co-carriage of specific HMRGs and ARGs.
Increasing concern over the therapeutic use of zinc in animal production potentially leading to an increased prevalence of LA-MRSA contributed to a phase-out of these products in the EU (Rensing et al., 2018). Therapeutic use of zinc was banned from June 2022 within the EU and whilst the UK was included with legislation passed before the UK left the EU, product still in date and remaining within the supply chain are permitted for use as part of the UK phase-out. Within the EU 27, zinc is now only permitted at concentrations up to 150 ppm for nutritional use, compared to concentrations of 2,500 ppm used previously for therapeutic use (Veterinary Medicines Directorate, 2022).
Two related studies by Agga et al. (2014, 2015) on the effects of copper supplementation (125 ppm vs 16.5 ppm) on AMR in weaned pigs used data from the same trial but analysed different parameters. The first study (Agga et al., 2014) found that copper MIC was not affected by copper supplementation or by pcoD gene carriage (a plasmid-borne copper HMRG). The second study (Agga et al., 2015) reported that copper supplementation was associated with a significant increase in tetP genes (which impart resistance to tetracyclines) among faecal E. coli but did not show a link with the pcoD gene (Agga et al., 2015). These studies observed that copper supplementation was associated with lowered blaCMY-2 gene copies from faecal E. coli. According to their results tetA and blaCMY-2 were positively associated with each other and negatively associated with both pcoD and tetB genes. They suggested that this points to the potential opportunity to select for a less harmful tetracycline resistance profile in E. coli by replacing in feed antibiotics by copper. A point highlighted in Van Noten et al.’s (2016) systematic review of this data. In their review of these data Wales & Davies (2015) concluded that it is possible that baseline levels of antibiotic and copper resistance/tolerance were sufficiently high in this study population that the copper supplementation was insufficient to select for reduced copper susceptibility or associated ARGs. Van Noten et al. (2016) judged the trials to be of intermediate methodological quality because of uncertainty concerning the independence of the samples (the same piglet could have been sampled at different weeks). We would agree that the data are not particularly compelling.
A Bavarian study observed that high concentrations of zinc and copper in pig manure (indicative of high concentrations in feed) may promote the spread of AMR of gut microbiota in pigs (Holzel et al. 2012). In the survey of manure samples from 305 pig farms the study found that supramedian concentrations of copper (388.5 ppm) and zinc (1,199.2 ppm) showed significant associations with the phenotypic antibiotic resistance of E. coli isolates in the manure. Bacterial resistance against ampicillin, augmentin (amoxicillin plus clavulanate), and piperacillin was significantly higher in E. coli from pig manure containing copper. While resistance rates against piperacillin and doxycycline in E. coli from pig manure were associated with zinc.
A possible effect of zinc feed supplementation on the mobility of ARGs in E. coli was observed by Bednorz et al. (2013), who reported an increased diversity of genotypes and plasmid profiles and increased MDR among weaning pigs supplemented with high concentrations of zinc (>2,000 ppm). The study found that 18.6% of the E. coli clones from the high zinc group were multi-resistant but not a single clone from the control group (50 to 70 ppm). An independent second study (Ciesinski et al., 2018) used the same feeding setup, but changed the experimental setup to focus on a complex analysis of resistance phenotypes rather than clonal diversity. They also observed that high dietary zinc feeding increased the proportion of MDR E. coli from weaned piglets, corroborating the finding of the previous study. The impact of zinc was observed in all three habitats tested (faeces, digesta, and mucosa). The authors suggested several possible mechanisms for their observations. One was co-selection, as some isolates had both zinc tolerance and antibiotic resistance. Another was enhanced exchange of MGEs under the influence of zinc. Differences in the plasmid profiles of clones of the zinc and control group were observed in the initial study (Bednorz et al., 2013).
In contrast, a further study by this group (Ghazisaeedi et al., 2020) argued against a co-selection mechanism of zinc and AMR suggesting that an explanation for an increase in MDR isolates from piglets with high zinc dietary feeding could be that antimicrobial-resistant bacteria are more tolerant to stresses such as zinc or copper exposure. In this further study, the group screened the phenotypic zinc/copper tolerance of 210 isolates (including antibiotic resistant, MDR, and non-resistant E. coli) selected from two, independent zinc-feeding animal trials. Importantly, no significant association was observed between AMR and phenotypic zinc/copper tolerance of the same isolates.
Medardus et al. (2014) also observed an effect of zinc as a feed supplement. They reported that among 349 Salmonella spp. isolates from nine pig units in the USA studied over a two-year period, an elevated zinc MIC was associated with the occurrence of the czcD-encoded zinc efflux pump but not with the concentration of faecal zinc. By contrast faecal copper concentration was associated with an elevated copper MIC, but not with the occurrence of a copper efflux gene pcoA. The same study reported that specific serovars were associated both with copper and zinc susceptibility and with patterns of antibiotic resistance; such resistances were not independently associated with copper or zinc susceptibility once serovar was taken into account. Concentrations of zinc and copper in the feed were between 79-7,384 ppm and 3-1,384 ppm, respectively.
In a study of faecal E. coli among 180 weaner pigs in the US, in-feed copper supplementation at a growth-promoting concentration (125 ppm) was associated with reduced susceptibility to chlortetracycline and oxytetracycline in E. coli (Shelton et al., 2009). No significant effects were observed for high concentrations (3,000 ppm) of added zinc.
Studies have reported conflicting evidence on an association between the supplementation of copper in feed of different animals and resistance in faecal enterococci. The development of tolerance to copper in enterococci is associated with the presence of tcrB, a copper HMRG, which is often located on a conjugative plasmid that may carry ARGs, thus contributing to co-selection (Yazdankhah et al., 2014). In a study of Enterococcus faecium isolated from pigs on Danish farms, tcrB was more frequently detected from the more intensively copper-supplemented livestock (Hasman & Aarestrup, 2002). Copper tolerance was strongly correlated with macrolide and glycopeptide resistance in isolates from pigs, and tcrB genes shown to be located on the same conjugative plasmid as ARGs erm(B) and vanA (associated with both vancomycin and teicoplanin resistance). In a further study by this group, weaner and grower pigs were given a heterogeneous inoculum of tcrB-positive and -negative E. faecium and reported that exposure at a commercial in-feed concentration of copper (175 ppm vs 6 ppm) was associated with a higher detection frequency of tcrB and of the linked erm(B) and vanA genes (Hasman et al., 2006). They also identified the tcr genes in the enterococcal species E. mundtii, E. casseliflavus, and E. gallinarum.
In contrast, two US studies by Amachawadi et al. (2010, 2015a) found no relationship between feeding weaned piglets with feed with elevated copper concentrations (125 ppm) compared to the control diet (16.5 ppm) and an increased prevalence of copper-resistant enterococci. Though one study by this team (Amachawadi et al., 2011), similar to the other trials, did show that elevated copper in feed could increase the prevalence of tcrB-positive enterococci. These studies did demonstrate a positive correlation between the presence of the tcrB gene and tolerance to copper and the possibility of transferring this gene to enterococci from the same and from different species (Amachawadi et al., 2010, 2011), but did not specifically examine AMR in these enterococci. A study by Ragland et al. (2006) reported no increase in vancomycin-resistant enterococci isolates from piglets (17 to 20 days old) receiving an increased copper or zinc supplementation (192.4 and 2,712.68 ppm, respectively) compared to the control group (11.2 and 120 ppm, respectively).
In a further study by Amachawadi et al. (2015b) copper fed to USA feedlot cattle at a growth promotion concentration (100 ppm) was observed to be associated with modest, but significantly increased frequencies (4.5% vs 2.0% in controls) of detection of tcrB-positive and macrolide-resistant erm(B)-positive E. faecium, whilst resistances to other screened antibiotics, including vancomycin, were unaffected (Amachawadi et al., 2015b), although an earlier study reported that feeding elevated copper (up to 100 ppm) and /or zinc (up to 300 ppm) to feedlot cattle had marginal effects on AMR of faecal E. coli (resistance to clindamycin, erythromycin, penicillin, tiamulin, tilmicosin, and tylosin) and enterococci [classified as susceptible or intermediate to chloramphenicol, ciprofloxacin, gentamicin, linezolid, penicillin, streptomycin, and vancomycin] (Jacob et al., 2010). In E. coli and Enterococcus spp., only minimal differences in MICs of copper, zinc, and antibiotics were noticed. The tcrB gene was not detected in faeces or in enterococcal isolates. The proportions of erm(B) and tet(M) were unaffected by copper or zinc supplementation although this was a relatively small-scale trial involving only 20 animals, with only 5 animals per treatment.
As previously mentioned, a study by Cufaoglu et al. (2022) investigated AMR and heavy metal tolerance (as well as biocide tolerance) amongst E. coli from chicken, cattle, and sheep in Turkey. A high prevalence of AMR (with 99% and 78% resistant to erythromycin and/or fosfomycin, respectively) were reported amongst isolates, along with a high prevalence of reduced susceptibility to zinc (62 %). Whilst HMRGs, BRGs, and ARGs were found in the same isolates, there were limited data presented to suggest that the use of heavy metals and/or biocides specifically were driving AMR.
A further study that undertook WGS of E. coli from veal calves found a higher proportion of AMR isolates with BRGs, sugE (80%), sugE1 (27%), and with 50% of isolates carrying the qacE∆1 gene (Haley et al., 2022). Furthermore, ARGs mph(A), dfrA17, aadA5, and blaCTX-M-15 were positively associated with silver (sil) and copper (pco) HMRGs. But a negative association was observed between HMRGs and some frequently identified ARGs (sul2, aph(3”)-Ib, and aph(6)-Id). The authors speculated that since copper is found in milk replacer and calf starter diets it may also be co-selecting for AMR, with some association between MGEs and HMRGs, BRGs, and ARGs.
A Chinese study (Yang et al., 2020a) examining E. coli and Salmonella spp. isolates from broiler farms and broiler meat found no association between ARGs and HMRG or BRGs in E. coli, but there was a positive association between HMRGs (including pcoR and zntA) and BRGs (sugE(c), emrE, mdfA, ydgE/ydgF, qacF, sugE(p) and qacE∆1). In Salmonella spp. isolates ARGs (including ß-lactam resistance genes (blaCTX, blaTEM and blaSHV), tetracycline resistance genes (tetA, tetB, and tetC) and sulfonamide resistance genes (sul1, sul2, and sul3)) were associated with HMRGs (including pcoR, pcoA, and pcoC), with some HMRGs (including pcoR and pcoA) associated with qacE∆1. No details of the Salmonella spp. were provided by the authors.
Whilst not evidence of co-selection, a novel genomic island (clusters of genes within a bacterial genome that appear to have been acquired by HGT) likely to be due to insertion of a plasmid was found in monophasic S. Typhimurium isolates from the UK and Italy during 2005-2012. The genomic island included a number of ARGs genes, but also gene clusters associated with tolerance to arsenic, cadmium, zinc and copper. These isolates formed a single clade (isolates composed of a common ancestor) distinct from recent monophasic epidemic clones previously described from North America and Spain. Furthermore, isolates within this clade had a significantly higher MIC for copper sulphate than those outside the clade and without the genomic island. The authors concluded that heavy metal supplements in feed within the gastrointestinal tract of pigs may have contributed to the success of this clade (Petrovska et al., 2016). This is also supported by work in the USA by Medardus et al. (2014) who also found a strong association between AMR and heavy metal tolerance among serotypes of Salmonella spp. of public health importance.
In two opinions, the EFSA FEEDAP Panel concluded that co-selection in the gut bacteria for tolerance to zinc and copper could not be excluded (EFSA FEEDAP Panel, 2014, 2016). While the opinion on zinc did not consider its impact on AMR in any detail, the opinion on copper in feed did consider its impact on AMR in detail, which was supported by a systematic literature review by Van Noten et al. (2016). While both of these opinions made recommendations (that were later actioned into regulations) for lower permitted concentrations of zinc and copper in animal feeds (as quoted in this report in a previous section) these concentrations were primarily based on dietary requirements rather than on any impact on co-selection risk.
Table 5: In-field studies that have addressed the impact of heavy metals on antibiotic resistance (AMR).
| Form of animal production | Context | Heavy metal | Species and strains | Impact on resistance | Stated antimicrobial resistance profiles (antimicrobial or class) | Antimicrobia I Resistance Gene (ARGs) present | Heavy metal Resistance Genes (HMRGs) present | Country | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Feedlot cattle | Feed supplementation | Copper, zinc | E.Coli, Enterococcus sp | No association with increased resistance | NS | erm(B), tet(M) | NS | USA | Jacob et al, 2010 |
| Weaning pigs | Therapy | Zinc | LA-MRSA | Association with increased resistance | Methicillin | mecA | czrC | Canada | Slifierz et al. 2015a |
| Weaning pigs | Therapy | Zinc | LA-MRSA | Association with increased resistance | Methicillin | mecA | czrC | Canada | Slifierz et al. 2015b |
| Weaned pigs | Feed supplementation | Copper | E.Coli | Association with change in resistance | NS | tetA, tetB, blaCMY-2 | pcoD | USA | Agga et al., 2014, 2015 |
| Weaning pigs | Feed supplementation | Copper, zinc | E.Coli | Copper associated with increased resistance, but not zinc | Chlortetracycline, Neomycin, Oxytetracycline, Tiamulin | NS | NS | USA | Shelton et al., 2009 |
| Weaning pigs | Feed supplementation | Zinc | E.Coli | Association with increased resistance | NS | blaTEM, tet(A), tet(B), sul1, sul2 | NS | Germany | Bednorz et al., 2013 |
| Pigs |
Therapy
|
Zinc | LA-MRSA | Association between zinc resistance gene and methicillin resistance. | Erythromycin, Penicillin, Tetracycline | mecA | czrC | Denmark | Aarestrup et al. 2010; Cavco et al. 2010 |
| Pigs | Feed supplementation | Copper | E.faecium | Association with increased resistance | Macrolides, Glycopeptide | erm(B), vanA | tcrB | Denmark | Hasman & Aarestrup, 2002 |
| Cattle | Feed supplementation | Copper | E. faecium, E. faecalis. | No association with increased resistance | NS | NS | tcrB | USA | Amachawadi et al., 2015a |
| Cattle | Feed supplementation | Copper | E. faecium | Association with increased resistance | Macrolides | erm(B) | tcrB | USA | Amachawadi et al., 2015b |
| Weaning pigs | Feed supplementation | Zinc | E.Coli | Association with increased resistance | NS | NS | NS | Germany | Ciesinski et al., 2018 |
| Weaning pigs | Feed supplementation | Zinc | E.Coli | Association with increased resistance, but no evidence of co-selection | β-lactamases (Ampicillin or cefotaxime), Tetracyclines (Tetracycline), Aminoglycosides (Streptomycin) and Potentiated Suphonamides (sulphamethoxazole/trimethoprim) | NS | NS | Germany | Ghazisaee di et al., 2020 |
As well as in feed, heavy metals may also be used as antimicrobial agents against multiple types of bacteria. Heavy metals, such as zinc and silver, are used for the treatment of burned skin surfaces, open wounds, and specific eye infections and have also been incorporated in medical devices (McDonnell & Russell, 1999; Maillard & Hartemann, 2012). AMR isolates of E. coli that also showed reduced susceptibility to silver (and copper) have been isolated from UK pig abattoirs, suggesting that co-selection is possible (Yang et al., 2020b). Few studies appear to have addressed whether their use as antimicrobial agents could be drivers for co-selection. Maillard & Hartemann (2012) called for a better understanding and control of silver usage to prevent its possible contribution to the spread of AMR. There is still clearly a need to evaluate the potential risk of the use of silver contributing to antibiotic resistance.
One of the few published studies to have considered co-selection (Elbehiry et al., 2019), demonstrated no cross-resistance between silver or gold tolerance (used in the form of nanoparticles) in adapted strains (previously subjected to sub-inhibitory treatments) of S. aureus associated with mastitis and isolated from dairy cattle and AMR. Gold nanoparticle treatments were observed to cause less development of resistance than silver treatments.
Copper and zinc are routinely used in the UK in antimicrobial footbaths in commercial dairy farming to prevent lameness caused by bacterial infections (Bell et al., 2014; Yu et al., 2017; Williams et al., 2019). Though there appears to be no evidence on their impact on co-selection. It is likely that their disposal into slurry tanks will lead to soil contamination and thus may be a driver for co-selection of AMR. This has been recognised (Williams et al., 2019) but does not appear to have been studied. Williams et al. (2019) estimated that nearly 400 million litres of cattle footbath waste is likely to be disposed annually into slurry tanks in the UK. They demonstrated that layered double hydroxides are effective in removing copper and zinc from a commercially available cattle footbath solution.
Few studies on the impact of heavy metals used in aquaculture on AMR were identified in our literature search. While there are studies on the impact of heavy metals in the environment on AMR in fish and seafood, these studies appear to relate mainly to the impact of pollution on wild caught species rather than on the impact of heavy metal use in aquaculture.
For example, a study of E. coli from pond sediment from fish farms in Nigeria showed co-occurrence of metal (copper and zinc) tolerance and antibiotic resistance (to β -lactams including 3rd-generation cephalosporins, the fluoroquinolones, potentiated sulphonamides, tetracyclines, aminoglycosides and phenicols) and a significant correlation between concentrations of these metals and AMR (Ajewole, 2021). There was an absence of detailed information from farms on the use of biocides or feed containing these metals to correlate use with development of AMR.
Studies have shown a correlation between heavy metal tolerance and AMR in bacteria associated with aquacultural environments. But no direct causative link between the use of heavy metals in feed or as an antifoulant have been made. Aeromonads and Pseudomonads from Australian rainbow trout and sediments displayed resistance to β-lactams, trimethoprim and florfenicol and reduced susceptibility to heavy metals [including zinc and copper] (Akinbowale et al., 2007). No link was made to any use of these heavy metals in aquaculture beyond speculation regarding the use of copper to control algae and parasites. Chenia & Jacobs (2017) observed a correlation between erythromycin resistance and copper tolerance in bacteria isolated from a tilapia aquaculture system, but while the authors suggested that copper use in feed and as an antifoulant could be responsible, this was not investigated. Neither of these studies investigated the presence of specific ARGs.
A study of the dissemination of resistance genes in duck/fish polyculture ponds, a typical farming model in some parts of China, showed significant correlations between concentrations of copper and zinc and numerous ARGs (Zhou et al., 2019). Concentrations of copper were significantly and positively correlated with the relative abundance of sul3, tetT, tetW, qnrB, qnrS, fexB, sul1, sul2, tetM, and qnrA genes. With zinc concentrations significantly correlated to relative abundance of sul2, sul3, tetM, tetA, tetT, tetW, qnrA, qnrB, qnrS, aac(6′)-Ib, qepA, blaSHV, cmlA, floR, fexA, cfr, and fexB genes. Again, while the authors suggested that differences in metal levels could be related to different feed formulations no levels of metals were measured in the feeds used.
A study by Romero et al. (2017) has been widely cited in the literature as providing evidence of the co-selection of heavy metals on AMR in seafoods. This study observed multiple tolerances (to biocides, heavy metals, and antibiotics) in 76% of isolates from a wide range of seafoods. ARGs detected included sul1 (43.33% of tested isolates), sul2 (6.66%), blaTEM (16.66%), blaCTX−M (16.66%), blaPSE (10.00%), blaIMP (3.33%), blaNDM−1 (3.33%), floR (16.66%), aadA1 (20.00%), and aac(6′)-Ib (16.66%). The copper HMRGs pcoA/copA, and pcoR were detected in 36.66% and 6.66% of selected isolates, respectively. These results suggest that exposure to heavy metals may co-select for AMR, although the fish and seafood sampled was purchased at a wide range of retail supermarkets and fish markets in the region of Jaen, Spain. While many samples were sea-caught fish; sea bass, salmon, and prawn samples were farmed and showed patterns of tolerance/resistance to heavy metals, and antibiotics. No direct comparison with any pattern of use of heavy metals during the husbandry of these seafoods could be made.
Overall, the literature on investigations of bacterial strains recovered from the field show evidence of associations/correlations between heavy metal use and increased resistance/tolerance to antimicrobial agents. Particularly there is evidence that high concentrations of copper or zinc may co-select AMR. This has led to a reduction in permitted concentrations in recent years. There is a need to establish whether current use (in feed and other uses) in food animal production in the UK represent a real hazard with respect to the selection of AMR.
Persistence of biocides and/or heavy metals in the environment
The environmental persistence of biocides depends on the nature, action, and use of the biocide. While non-oxidising biocides (such as QACs) are likely to persist in the environment (Wales & Davies, 2015; Mulder et al., 2018; Davies & Wales, 2019), oxidising agents, (such as ozone, hydrogen peroxide, chlorine dioxide, sodium hypochlorite, peracetic acid and iodophors) by their nature are unstable and prone to degradation and rapidly breakdown during use. While several reviews (Capita & Alonso-Calleja, 2013; Wales & Davies, 2015; Davies & Wales, 2019) express concern regarding the persistence of biocides, such as QACs, in the environment they cite no specific studies that appear to have studied this or provide evidence exactly of how long biocides, such as QACs, may persist in the environment. Hegstad et al. (2010) cite several studies on the fate of a QAC in the environment, one of which reported a biodegradation of 36% in 28 days, but there does not appear to be any studies that have looked at the fate and persistence of on-farm biocides in-field. A comprehensive review of predicted and measured concentrations and fate of QACs in soils and their implications on AMR development was undertaken by Mulder et al. (2018). They predicted that concentrations of QACs in manure-amended soils could reach 3.5 ppm after 1 year, assuming zero biodegradation, but highlighted the lack of data on this and whether QACs could accumulate in soil over time.
While many biocides breakdown during use, heavy metals do not biodegrade, are very persistent and will accumulate in the environment. Heavy Metals, along with antimicrobial-resistant bacteria, ARGs, and HMRGs, may be introduced into soil and water through sewage systems, direct excretion, land application of biosolids or animal manures as fertilisers, and irrigation with wastewater or treated effluents (Yazdankhah et al., 2018). In England and Wales, food animal production has been estimated to be a major source of environmental contamination by zinc and copper (Nicholson et al., 1999, 2003, 2006). Livestock manure was found to be responsible for an estimated 37%–40% of total zinc and copper inputs. Denmark has maintained a national monitoring program of heavy metals in the environment for the last 28 years to better understand the effects of these practices on the environment. The values and analyses published in 2016 indicate that the use of pig slurry has led to a significant increase in the measured concentrations of copper and zinc in soil (Rensing et al., 2018). The persistence of heavy metals in agricultural soil may lead to leaching into natural water thus impacting on irrigation and aquaculture.
A recent EFSA report highlighted that further research is required to quantify the concentrations of potentially co-selective residues of biocides and heavy metals in manures, agricultural, and aquaculture environments to facilitate risk assessment of the role that they may play in co-selection for AMR (EFSA BIOHAZ Panel, 2021).
Dissemination of AMR from animal manures to agricultural soils
Land application of animal manure is a common agricultural practice potentially leading to dispersal and propagation of ARGs in environmental settings. The dissemination of antimicrobial-resistant bacteria and ARGs from animal manure and slurry to agricultural soils has been addressed in numerous studies and reviews. It was not the purpose of this study to review this evidence, only the specific evidence on the impact of biocides and/or certain heavy metals used in animal production on AMR in this context. There is clear evidence that agricultural soils are a vast reservoir of antimicrobial-resistant bacteria and ARGs, and that the application of animal manure and/or slurry contributes to this reservoir. Overall, environmental factors can have a high impact on selective pressures, distribution, and diversity of AMR in agricultural soil. Namely, soil characteristics, such as silt, clay, organic matter, and pH, have been shown to correlate with the relative abundance of antimicrobial-resistant bacteria and ARGs (Wang et al., 2022a). This has been reviewed by Zalewska et al. (2021) and Wang et al. (2022a). Different manure sources may influence the fate of resistome in agri-ecosystems with studies demonstrating that the application of pig and poultry manures leading to a greater abundance of ARGs than cattle manure (Zhang et al., 2017b; Duan et al., 2019).
There have been numerous studies on how animal manure may be treated or processed to reduce the transmission of antimicrobial-resistant bacteria and ARGs into the environment. These methods have been reviewed by Liu et al. (2021a), Ezugworie et al. (2021), EFSA BIOHAZ Panel (2021), Wang et al. (2022a, b), amongst others. Commonly used methods include aerobic composting, anaerobic digestion, and aerobic digestion. Other alternative methods include the use of biochar, nano-materials, and phage (though, as previously discussed, phage have also been implicated in the transfer of resistance genes within the soil microbiota, albeit experimentally). Ezugworie et al. (2021) concluded that no single composting protocol completely eliminated ARGs and that a combination of protocols could yield better results. Available data indicate that none of these methods are effective at eliminating antimicrobial-resistant bacteria and ARGs. It is clear that additional research is needed to determine optimum methods in a UK context for reducing/eliminating antimicrobial-resistant bacteria and ARGs from stored manure prior to use in the environment. A recent EFSA report also highlighted that such measures may increase storage and equipment resources requirements and may reduce the fertiliser value (EFSA BIOHAZ Panel, 2021), although the report did not cite specific evidence in relation to this conjecture.
There is some evidence that a delay between the application of manure and plant life cycle (germination, growth, or harvest) of crops may reduce contamination and internalisation with antimicrobial-resistant bacteria and ARGs (EFSA BIOHAZ Panel, 2021). According to a recent EFSA BIOHAZ Panel report (EFSA BIOHAZ Panel, 2021) further research is required to define what a suitable delay may be.
Impact of biocides on AMR in animal manures and agricultural soils
No evidence has been found in the literature on the impact of biocides used in food animal production on antimicrobial-resistant bacteria or genes detected in manure and manure enriched soils. A review on the occurrence of biocides in 2016 by Wohde et al. (2016) cited only three studies at the time on the occurrence of biocides in manure. The studies cited were mainly on methods of detection and contained no evidence on what biocides may persist in animal manure. Wohde et al. (2016) at the time highlighted that the occurrence of biocides in manure had been neglected, which would appear to still be the case.
Impact of heavy metals on AMR in animal waste and agricultural soils
Due to negligible absorption in the gut, high concentrations of zinc and copper in feed results in high concentrations of these metals in animal waste. In the United Kingdom, Nicholson et al. (1999) analysed manure samples from commercial farms in England and Wales, finding zinc concentrations in 12 samples ranging from < 5 to 2500 ppm with typical concentrations of approximately 500 ppm. Data collected in China (Table 6) report that concentrations of copper and zinc are higher in pig manure than other animal manures (Wang et al., 2013). This is likely to be the case in the UK, but detailed data are lacking. It is also likely that reductions in the concentrations of heavy metals permitted in food animal production in the UK (as previously discussed in this report) may have reduced the concentration of these metals in animal waste, but again data are lacking. The environment close to aquaculture production sites have also been reported to contain elevated concentrations of copper and zinc due to the added minerals in fish feed (Burridge et al., 2010). We have found no clear evidence in the literature linking copper and zinc concentration in fish feed used in aquaculture to levels at production sites, whether in the water or sediment. Heavy metal concentrations in water, sediment, soil, and manure reported in the literature were compiled by Seiler & Berendonk in 2012. They introduced the notion that concentrations of heavy metals may need to accumulate to critical concentration before they can trigger co-selection of AMR. As also highlighted by the FAO and WHO (FAO/WHO, 2019) and Arya et al. (2021), there are little data on what these threshold values should be in order to inform suitable standards for metal concentrations in food animal production. Arya et al. (2021) developed a general model to provide a general mechanistic framework to predict minimal co-selective concentrations for metals, based on knowledge of their toxicity at different concentrations. They predicted MSCs of 5.5, 1.6, and 0.152 mg/L for copper, zinc, and silver, respectively. Comparing these thresholds with metal concentrations from slurry and slurry-amended soil from a UK dairy farm that used copper and zinc as additives for feed and in an antimicrobial footbath (at current permitted concentrations) they predicted that the slurry (which contained 22.31 and 32.16 mg/L of copper and zinc, respectively) would be co-selective, but not the slurry-amended soil (which contained only 0.068 and 0.16 mg/L of copper and zinc, respectively).
Table 6: Heavy metal contents (mean (standard deviation) ppm) reported in animal feed and manures (adapted from Wang et al., 2013).
| Animal type | Copper content in feed | Zinc content in feed | Copper content in manure | Zinc content in manure |
|---|---|---|---|---|
| Pig | 82.0 (87.7) | 149.2 (280.1) | 288.6 (382.3) | 599.1 (1194.4) |
| Sow | 42.9 (66.9) | 82.2 (48.5) | 136.4 (193.3) | 483.5 (522.7) |
| Cattle (dairy) | 19.8 (13.1) | 98.6 (78.2) | 56.1 (51.7) | 212.6 (103.3) |
| Poultry | 30.4 (41.4) | 102.6 (46.1) | 141.7 (265.1) | 432.3 (287.1) |
| Poultry (broiler) | 49.0 (57.0) | 111.2 (55.0) | 144.3 (208.5) | 351.6 (198.2) |
Studies (examples being Ji et al., 2012; Zhou et al., 2016; Peng et al., 2017; Guo et al., 2018; Duan et al., 2019; Wu et al., 2020; Dong et al. 2021; Liu et al., 2021a; Peng et al., 2021; Xue et al., 2021; Mazhar et al., 2021; Liu et al., 2022) have shown a positive correlation between the presence of heavy metals in animal manure and in agricultural soils enriched with animal manure containing heavy metals. The specific source of these heavy metals was not identified. While feed and/or medicines are often cited as probable sources of these metals, studies fail to provide clear evidence of a correlation between concentrations of these metals existing in feed and/or medicines and corresponding levels in manure and manure-enriched soils. Thus, there is no firm evidence of a causative relation in this matter. Microbial communities may be shaped by exposure to different agents and furthermore heavy metals within natural environments have been shown to significantly impact on the structure of microbial communities (Li et al, 2020). There is also some evidence that presence of heavy metals may have a positive effect on the HGT potential of ARGs in soil (Peng et al., 2017; Tongyi et al., 2020; Yuan et al., 2020; Liu et al., 2021b; Li et al., 2022c). Though how heavy metals enhance the mechanism of genetic transfer is not clear, although there is some evidence that livestock-associated bacteria may carry more MGEs than bacteria from other sources, such as clinical environments (Yuan et al., 2020) and manure has been shown to be rich in MGEs carrying HMRGs co-occurring with ARGs (Liu et al., 2022), indicating the importance of MGEs mediating in co-selection. Positive correlations between the presence of heavy metals and ARGs/MGEs in manures and enriched soils have been observed (Peng et al., 2021; Tu et al., 2023). A positive correlation between concentrations of copper and zinc and the integron-integrase gene, intI1, (linked to genes conferring resistance/tolerance to antibiotics, biocides, and heavy metals) has been observed in soil continuously amended with manure for 30 years (Peng et al., 2017). Although this particular study observed that heavy metals, and the source of manure influenced ARGs in the soil, it did not directly trace the source of these heavy metals to use in feed, although feed was suggested as a likely source. A recent study by Li et al. (2022a) suggests that while low levels of copper and zinc in pig manure may alter resistance and MGE compositions they may not be the primary drive for ARG transmission. The type of soil, which as discussed may impact on AMR, is often not stated in the literature (Wang et al., 2022a). It should also be noted that these studies have been carried out in China, where different production regimes may be practiced than in the UK (including the more widespread use of antibiotics) and there appears to be little evidence regarding the impact of European food animal production practices on ARGs in soil. The studies also lack comparison with control farms with no use of metals and/or antibiotics which would allow strong conclusions regarding the use of heavy metals.
Since heavy metals are non-biodegradable, they may be a continuous pressure on co-selection of AMR during the composting of animal manure and waste bedding (Li et al., 2015; Wang et al., 2019; Liu et al., 2021a; Ezugworie et al., 2021). Limits on heavy metals concentrations in compost have been issued by different countries, limits for copper and zinc are shown in Table 7.
Table 7: Copper and zinc limits (ppm) in compost permitted by different countries (adapted from Wang et al., 2019).
| Heavy metal | Austria | Belgium | Switzerland | Germany | Italy | Spain | Canada |
|---|---|---|---|---|---|---|---|
| Copper (ppm) | 400 | 100 | 150 | 100 | 300 | 1750 | 100 |
| Zinc (ppm) | 1000 | 1000 | 500 | 400 | 500 | 4000 | 500 |
Some studies have investigated the impact of biochar in mitigating the impact of the presence of heavy metals in animal manure on AMR during composting or digestion, or in manure-enriched soil (Liu et al., 2021a, Wang et al., 2022a; Ejileugha, 2022). Biochar has been reported to reduce the bioavailability of heavy metals, prevent HGT, and eliminate ARGs carried by MGEs (Ejileugha, 2022; Wang et al., 2022a). More novel mitigation treatments for removing heavy metals include electro remediation, which passes an electric current through liquid manure and metal ions are precipitated on an electrode (Hejna et al., 2018). At present the technology is unproven at the farm-scale and is unlikely to be cost-effective.
AMR transfer from soil to crops and foods of plant origin
The dissemination of antimicrobial-resistant bacteria and genes from manure to agricultural soils to crops and foods of plant origin has been addressed in numerous studies and reviews (Hölzel et al. 2018; FAO/WHO, 2019; Zalewska et al. 2021; EFSA BIOHAZ Panel, 2021; Wang et al. (2022a), amongst others). A number of reviews (including recent reviews by FAO/WHO, 2019 and EFSA BIOHAZ Panel, 2021) have cited evidence that antimicrobial-resistant bacteria and ARGs from manure-amended soils can potentially disseminate from soil microbiota to plant microbiota, thus may be an important route for AMR transmission in foods of plant origin. Since fruits and vegetables are frequently eaten raw or with minimal processing they can potentially serve as a source of dietary exposure to antimicrobial-resistant bacteria and ARGs of animal-origin.
It was not the purpose of this study to review this evidence, only the evidence on the impact of biocides and/or certain heavy metals used in animal production on AMR. The FAO/WHO (2019) report also suggested that heavy metals (for example, copper and zinc in feed) should be considered in terms of probability of selection.
While studies, such as Buta et al. (2021), have observed an association between the presence of heavy metals and ARGs in animal manure which migrated with the manure to agricultural soils enriched with animal manure and these to crops, a causal link to the use of heavy metals in animal production is not identified. Longitudinal studies on the impact of biocides and/or heavy metals during animal production on the transmission of AMR to crops and foods of plant origin are lacking.
Overall, there appears to be little compelling evidence in the literature on this subject and a better understanding is needed on how ARGs may transfer from the soil to plants and the risk of ARGs to humans from consumption of plants containing antimicrobial-resistant bacteria and/or ARGs, particularly those intended to be consumed raw.
AMR transfer from soil to animals and foods of animal origin
While faeces, fertilisers of animal origin (for example, manure and slurry), and bedding have been identified as potential transmission routes to the dissemination of antimicrobial-resistant bacteria and genes in animals and foods of animal origin there is little information on their importance (EFSA BIOHAZ Panel, 2021). Longitudinal studies on the impact of biocides and/or heavy metals during animal production on the transmission of AMR to animals and foods of animal origin are lacking. While there is evidence of co-carriage of BRGs/HMRGs and ARGs in retail meats (for example Zou et al. 2014 and Yang et al., 2020a) which is of concern, studies showing a clear relationship between the use of biocides and/or heavy metals on the farm contributing to AMR in retail meats is lacking.
Several uncertainties have been identified related to the probability of co-selection of, and transmission of, AMR due to the use of biocides and heavy metals in food animal production. Many of these are caused by knowledge and data gaps, and the limitations of the published studies identified and reviewed. Such gaps have also been identified in numerous other reviews, including those listed in Table 4, and SCENIHR (2010) amongst others. Conclusive evidence of the impact of the use of biocides and heavy metals in food animal production on AMR cannot be established because of the high level of uncertainty in the literature. As highlighted in other similar reviews (such as SCENIHR, 2010; Davies & Wales, 2019; FAO/WHO, 2019; EFSA BIOHAZ Panel, 2021) it is clear that more robust experimental strategies and testing are needed to identify risk. Intensive agricultural activities are likely to contribute to the selection and development of AMR but further evidence, particularly in the UK context, is needed.
Overall, there is a particular lack of evidence on how biocides and heavy metals may impact on AMR in aquaculture that needs to be addressed. It is likely that much of the work done in the context of general terrestrial systems will not apply to aquaculture where environmental conditions are different and both bacteria and agents likely to behave differently. The impact of different aquaculture systems, for example marine or land based Recirculating Aquaculture Systems (RAS), need to be addressed.
There is compelling theoretical and laboratory experimental evidence that certain biocides may co-select for AMR. Studies that have investigated the effects of biocides on the development of AMR are predominantly laboratory-based, and translation to real-world environmental conditions is both difficult and limited. Longitudinal studies on the impact of biocides during animal production on the co-selection and transmission of AMR to animals and foods of plant and animal origin are lacking.
It is frequently cited in the literature that some biocides may leave residues that could co-select for AMR (FAO/WHO, 2019). Therefore, such biocides may be less appropriate for use in food animal production due to their potential for impact on the environment and the potential emergence of resistance. It is unclear in the literature what specific biocides may present such a risk and there are no data on whether biocides used in food animal production persist in the food animal production environment. It is unclear how long biocides can persist in the environment, at what concentration, and especially what (minimum or threshold) concentration can trigger AMR selection, and how this may be affected by environmental conditions.
Within the UK context there are insufficient data about the amounts and types of biocides used in terrestrial food animal production and aquaculture. This is important for informing future studies in the field or laboratory to provide robust data to assess the risk of such products on AMR selection.
There is compelling evidence that heavy metals will persist, accumulate, and may impact on the development of AMR in animal production environments for many years. Longitudinal studies on the impact of heavy metals during animal production on the co-selection and transmission of AMR to animals and foods of plant and animal origin are lacking. Permitted concentrations of zinc and copper used in food animal production have been reduced in recent years in the UK. It is unclear as to whether this has reduced the risk of these metals driving the selection and transmission of AMR.
There is evidence that levels of biocides and heavy metals need to accumulate to critical concentrations (MSCs) before they can trigger co-selection of AMR. As highlighted by the FAO and WHO (FAO/WHO, 2019) and Arya et al. (2021), there are little data on what these threshold values should be in order to inform suitable standards for biocide/heavy metal concentrations in food animal production.
It has been theorised that heavy metals in certain forms (as stable metal compounds that do not release free metal ions) may provide nutrition to food-producing animals but not be toxic to bacteria, and hence their use in feed would not co-select for AMR in bacteria (Yu et al., 2017). There does not appear to be any published evidence supporting this hypothesis.
A further issue is that many studies when examining isolates apply different methodologies and criteria for determining resistance/tolerance to biocides and heavy metals. There is a clear need for standardisation of methodology for both examining isolates and for also determining the concentration of agents within samples and concordance of resistance/tolerance phenotype with BRGs and HMRGs. A greater understanding of the nature and behaviour of biofilms in the field on the persistence and transmission of AMR is also required. The effects of field biocides on MGEs (such as plasmids) carrying resistance genes and the potential for HGT from MGEs that may persist following cleaning and disinfection also need to be explored.
Furthermore, many studies focus on a narrow range of bacteria and ignore the wider microbial community and upon which heavy metal and biocides could be exerting co-selection of AMR within the community or facilitating transfer of AMR via HGT.
Mitigation strategies
As Singer et al. (2016) stated “[the] understanding of AMR in the environment is so unsatisfactory that there is very little that can be suggested for mitigation without employing the precautionary principle as the primary rationale for action”. This remains the case and we would echo that statement. A better understanding of how biocides and/or heavy metals can influence AMR and ARGs in the primary food production environment is needed to inform the development of effective mitigation measures. As highlighted by Maillard (2020), and others, there is a conundrum that while the use of biocides is important to control microbial pathogens inappropriate applications may lead to an exacerbation of AMR. Similarly, the use of heavy metals in food animal production provide benefits (particularly to animal health), but may contribute to the spread of AMR.
Overall, when considering the use of biocides and heavy metals in food animal production and mitigation strategies, a clear distinction should be made between the risk of acquired antibiotic cross-resistance and co-selection through the use of sub-inhibitory supplementary concentrations of heavy metals and biocide and their use at concentrations that kill or inactivate. While biocides are in the main applied at killing or inactivation concentrations, some may be used in feed. In the main heavy metals are used at sub-inhibitory supplementary concentrations and will be present in the environment at these concentrations, and it is possible that some may be used at killing or inactivation concentrations in medical treatments.
Use of biocides in food animal production
Despite current gaps in knowledge, action can be taken to mitigate potential risks by providing clear guidance to manufacturers and users of biocidal products on practices aimed at minimising the potential development of resistance. For example, the appropriate use of biocides in keeping with the manufacturer’s instructions and the intended product use, and validation of effectiveness specific to the application. Improper or excessive use of biocides, and use at sub-inhibitory concentrations in the food production chain should be avoided as it may drive the emergence of AMR. It would be prudent for manufacturers of biocides used in food animal production to investigate whether cross-resistance to clinically-important antimicrobials is likely to occur under conditions of prescribed use.
Use of heavy metals in food animal production
While there are gaps in knowledge, effective strategies should focus on a reduction in the heavy metal input/output ratio in livestock and aquatic farmed animals. Recent reductions in permitted concentrations and a ban on therapeutic use are mitigation strategies that are already in place and should reduce risk. There is still a need to establish whether current concentrations in food animal production in the UK represent any real hazard with respect to the selection of AMR. Formulated diets that increase the efficiency of nutrient retention by animals, decrease their excretion in faeces and urine and reduce the import of nutrients in feed and mineral mixtures are still important mitigating strategies. Different approaches to reduce the content/impact of heavy metals in manure/slurry have been studied but optimum mitigation strategies not yet confirmed. Additional research is needed to determine optimum methods in a UK context for reducing/eliminating heavy metals and antimicrobial-resistant bacteria, ARGs, and HMRGs from stored manure/slurry prior to use in the environment. A recent EFSA report also highlighted that such measures may increase storage and equipment resources requirements and may reduce the fertiliser value of the manure/slurry (EFSA BIOHAZ Panel, 2021).
Conclusions
On reviewing the published evidence, our conclusions regarding the project’s four key questions (terms of reference) are thus:
Is there evidence to show that biocides and heavy metals used in food animal production have an impact on the development of AMR?
We have found that there is some evidence that biocides and heavy metals used in food animal production may have an impact on the development of AMR and either resulting in reduced susceptibility to drugs or clinically significant resistance. There is more compelling evidence regarding the use of heavy metals than there is on the use of biocides.
How long are biocides and heavy metals able to persist in animal production environments and how does this impact on the development of AMR and associated risks?
There is evidence that heavy metals will persist, accumulate, and may impact on the development of AMR in animal production environments for many years. There is less evidence on the persistence and impact of biocides. There is some evidence that while many biocides will rapidly break-down in the environment, some, such as QACs, may persist. There is little evidence on how long this persistence may be in animal production environments, and what the impact on AMR may be.
What evidence is there that biocide and heavy metal associated AMR enters the food chain through products of animal origin or as a result of crop contamination?
There is no clear evidence of biocide and/or heavy metal associated AMR entering the food chain, through products of animal origin or as a result of crop contamination due to their use in food animal production. Published studies that have demonstrated an association between biocide and/or heavy metal use and increased AMR/ reduced susceptibility risk in live animals, manure, slurry, or soil have not looked beyond these points at how this use may impact on AMR risk in food. Although there is evidence of the co-carriage of BRGs/HMRGs and ARGs in retail meats.
Is there a potential risk to the consumer from AMR acquired through the use of biocides and heavy metals in food animal production?
It is recognised that AMR in food is a risk to consumer health, and that food animal production has an impact on this risk. While there is certainly a theoretical risk, we have found no published evidence that has specifically demonstrated that the use of biocides and/or heavy metals in food animal production increases the risk of the consumer acquiring antimicrobial-resistant bacteria from food or has quantified that risk. There does not appear to be sufficient evidence to undertake such an assessment of risk.
A central question was whether the release of chemicals like biocides (in particular disinfectants) and/or heavy metals from food animal production has the potential to create local concentrations where AMR can emerge and spread (as bacteria or genes) and whether this presents a potential risk to the consumer as a result. In our opinion there does appear to be sufficient evidence that that this is possible and that there is a potential risk to the consumer. Nevertheless, there do not appear to be sufficient data to undertake such an assessment of risk and focussed in-field studies need to be carried out to fill this evidence gap and provide the data required to assess this risk.
Recommendations
The NAP highlighted that this subject (the role of heavy metals and other biocides in the environment in promoting AMR) remains an evolving area of research that requires further study. This remains the case.
While recognising that similar recommendations have been in past reviews of this subject (Table 4). In our opinion the following studies are required to improve our understanding of the role of biocides and heavy metals used in food animal production have in promoting AMR:
- To determine the amounts, types, and concentrations of biocides used in terrestrial food animal production and aquaculture in the UK and whether biocides persist in these environments, and at what concentrations.
- Implementing the monitoring of copper and zinc pollution from agriculture in areas in which food-producing animals are fed (including aquaculture), with particular attention to the potential development of microbial AMR in the environment. The data would help to identify any area under risk.
- Monitoring the occurrence of biocide/heavy metal tolerance and cross- and co-resistance in bacteria isolated from terrestrial food animal production and aquaculture environments in the UK.
- To determine the thresholds for biocide and heavy metal concentrations (MSCs) that could co-select for AMR in the animal production environment (including the aquatic environment), animal waste, manure, and manure enriched soils.
- To determine optimum methods in a UK context for reducing/eliminating heavy metals and antimicrobial-resistant bacteria, ARGs, and HMRGs from stored manure/slurry prior to use in the environment.
- To access through controlled longitudinal in-field trials whether there is a causal impact on the use of biocides and/or heavy metals used, at different concentrations, in food animal production (land and aquatic), impact on AMR selection and transmission in food animals and transfer to foods of animal origin, and impact on AMR selection and transmission in agricultural soil and subsequent transfer to crops and foods of plant origin to determine the risk.
- Applying culture independent approaches, such as shotgun metagenomics and exploring metagenomes, to examine samples and explore correlation more widely in less well characterised bacterial communities for BRGs/HMRGs and ARGs.
Aarestrup, F. M., Cavaco, L., & Hasman, H. (2010). Decreased susceptibility to zinc chloride is associated with methicillin resistant Staphylococcus aureus CC398 in Danish swine. Veterinary Microbiology, 142(3-4), 455-457. https://doi.org/10.1016/j.vetmic.2009.10.021
Abd El-Aziz, N. K., Ammar, A. M., El Damaty, H. M., Abd Elkader, R. A., Saad, H. A., El-Kazzaz, W., & Khalifa, E. (2021). Environmental Streptococcus uberis associated with clinical mastitis in dairy cows: virulence traits, antimicrobial and biocide resistance, and epidemiological typing. Animals, 11(7), 1849. https://doi.org/10.3390/ani11071849
Agga, G. E., Scott, H. M., Amachawadi, R. G., Nagaraja, T. G., Vinasco, J., Bai, J., Norby, B., Renter, D. G., Dritz, S. S., Nelssen, J. L., & Tokach, M. D. (2014). Effects of chlortetracycline and copper supplementation on antimicrobial resistance of fecal Escherichia coli from weaned pigs. Preventive Veterinary Medicine, 114(3-4), 231-246. http://dx.doi.org/10.1016/j.prevetmed.2014.02.010
Agga, G. E., Scott, H. M., Vinasco, J., Nagaraja, T. G., Amachawadi, R. G., Bai, J., Norby, B., Renter, D. G., Dritz, S. S., Nelssen, J. L., & Tokach, M. D. (2015). Effects of chlortetracycline and copper supplementation on the prevalence, distribution, and quantity of antimicrobial resistance genes in the fecal metagenome of weaned pigs. Preventive Veterinary Medicine, 119(3-4), 179-189. http://dx.doi.org/10.1016/j.prevetmed.2015.02.008
Ajewole, O. A., Ikhimiukor, O. O., & Adelowo, O. O. (2021). Heavy metals (Cu and Zn) contamination of pond sediment and co-occurrence of metal and antibiotic resistance in Escherichia coli from Nigerian aquaculture. International Journal of Environmental Studies, 78(5), 773-784. https://doi.org/10.1080/00207233.2020.1804741
Akinbowale, O. L., Peng, H., Grant, P., & Barton, M. D. (2007). Antibiotic and heavy metal resistance in motile aeromonads and pseudomonads from rainbow trout (Oncorhynchus mykiss) farms in Australia. International Journal of Antimicrobial Agents, 30(2), 177-182. https://doi.org/10.1016/j.ijantimicag.2007.03.012
Al-Mnaser, A. A., & Woodward, M. J. (2020). Sub-lethal concentrations of phytochemicals (carvacrol and oregano) select for reduced susceptibility mutants of O23: H52 Polish Journal of Microbiology, 69(1), 121-125. https://doi.org/10.33073/pjm-2020-003
Alderton, I., Palmer, B. R., Heinemann, J. A., Pattis, I., Weaver, L., Gutiérrez-Ginés, M. J., Horswell, J., & Tremblay, L. A. (2021). The role of emerging organic contaminants in the development of antimicrobial resistance. Emerging Contaminants, 7, 160-171. https://doi.org/10.1016/j.emcon.2021.07.001
Allen, K. J., Wałecka-Zacharska, E., Chen, J. C., Katarzyna, K. P., Devlieghere, F., Van Meervenne, E., Osek, J., Wieczorek, K., & Bania, J. (2016). Listeria monocytogenes–An examination of food chain factors potentially contributing to antimicrobial resistance. Food Microbiology, 54, 178-189. https://doi.org/10.1016/j.fm.2014.08.006
Alonso-Hernando, A., Capita, R., Prieto, M., & Alonso-Calleja, C. (2009). Comparison of antibiotic resistance patterns in Listeria monocytogenes and Salmonella enterica strains pre-exposed and exposed to poultry decontaminants. Food Control, 20, 1108–1111. https://doi.org/10.1016/j.foodcont.2009.02.011
Álvarez-Martínez, F. J., Barrajón-Catalán, E., Herranz-López, M., & Micol, V. (2021). Antibacterial plant compounds, extracts and essential oils: An updated review on their effects and putative mechanisms of action. Phytomedicine, 90, 153626. https://doi.org/10.1016/j.phymed.2021.153626
Amachawadi, R. G., Scott, H. M., Aperce, C., Vinasco, J., Drouillard, J. S., & Nagaraja, T. G. (2015b). Effects of in-feed copper and tylosin supplementations on copper and antimicrobial resistance in faecal enterococci of feedlot cattle. Journal of Applied Microbiology, 118, 1287–1297. https://doi.org/10.1111/jam.12790
Amachawadi, R. G., Scott, H. M., Vinasco, J., Tokach, M. D., Dritz, S. S., Nelssen, J. L., & Nagaraja, T. G. (2015a). Effects of in-feed copper, chlortetracycline, and tylosin on the prevalence of transferable copper resistance gene, tcrB, among fecal enterococci of weaned piglets. Foodborne Pathogens and Disease, 12(8), 670-678. https://doi.org/10.1089/fpd.2015.1961
Amachawadi, R. G., Shelton, N. W., Jacob, M. E., Shi, X., Narayanan, S. K., Zurek, L., Dritz, S. S., Nelssen, J. L., Tokach, M. D., & Nagaraja, T. G. (2010). Occurrence of tcrB, a transferable copper resistance gene, in fecal enterococci of swine. Foodborne Pathogens and Disease, 7(9), 1089-1097. https://doi.org/10.1089/fpd.2010.0540
Amachawadi, R. G., Shelton, N. W., Shi, X., Vinasco, J., Dritz, S. S., Tokach, M. D., Nelssen, J. L., Scott, H. M., & Nagaraja, T. G. (2011). Selection of fecal enterococci exhibiting tcrB-mediated copper resistance in pigs fed diets supplemented with copper. Applied and Environmental Microbiology, 77(16), 5597-5603. https://doi.org/10.1128/AEM.00364-11
Amjad, Z. (2010). The science and technology of industrial water treatment. CRC press. https://doi.org/10.1201/9781420071450
Anand, T., Bera, B. C., Vaid, R. K., Barua, S., Riyesh, T., Virmani, N., Hussain, M., Singh, R. K., and Tripathi, B. N. (2016). Abundance of antibiotic resistance genes in environmental bacteriophages. Journal of General Virology, 97(12), 3458–66. https://doi.org/10.1099/jgv.0.000639
Anedda, E., Farrell, M. L., Morris, D., & Burgess, C. M. (2023). Evaluating the impact of heavy metals on antimicrobial resistance in the primary food production environment: A scoping review. Environmental Pollution, 121035. https://doi.org/10.1016/j.envpol.2023.121035
Aperce, C. C., Amachawadi, R., Van Bibber-Krueger, C. L., Nagaraja, T. G., Scott, H. M., Vinasco-Torre, J., & Drouillard, J. S. (2016). Effects of menthol supplementation in feedlot cattle diets on the fecal prevalence of antimicrobial-resistant Escherichia coli. PloS one, 11(12), e0168983. https://doi.org/10.1371/journal.pone.0168983
Argudín, M. A., Lauzat, B., Kraushaar, B., Alba, P., Agerso, Y., Cavaco, L., Butaye, P., Concepción Porrero, M., Battisti, A., Tenhagen, B.-A., Fetsch, A., & Guerra, B. (2016). Heavy metal and disinfectant resistance genes among livestock-associated methicillin-resistant Staphylococcus aureus isolates. Veterinary Microbiology, 191, 88-95. https://doi.org/10.1016/j.vetmic.2016.06.004
Arya, S., Williams, A., Reina, S. V., Knapp, C. W., Kreft, J. U., Hobman, J. L., & Stekel, D. J. (2021). Towards a general model for predicting minimal metal concentrations co-selecting for antibiotic resistance plasmids. Environmental Pollution, 275, 116602. https://doi.org/10.1016/j.envpol.2021.116602
Bassani, J., Paravisi, M., Wilsmann, D. E., Borges, K. A., Furian, T. Q., Salle, C. T., Moraes, L. S., & Nascimento, V. P. (2021). Antimicrobial and disinfectant resistance of Salmonella Heidelberg from Brazilian flocks did not increase for ten years (2006-2016). Pesquisa Veterinária Brasileira, 41, e06818. https://doi.org/10.1590/1678-5150-PVB-6818
Becerril, R., Nerin, C., & Gomez-Lus, R. (2012). Evaluation of bacterial resistance to essential oils and antibiotics after exposure to oregano and cinnamon essential oils. Foodborne Pathogens and Disease, 9, 699–705. https://doi.org/10.1089/fpd.2011.1097
Bednorz, C., Oelgeschlaeger, K., Kinnemann, B., Hartmann, S., Neumann, K., Pieper, R., Bethe, A., Semmler, T., Tedin, K., Schierack, P., Wieler, L. H., & Guenther, S. (2013). The broader context of antibiotic resistance: Zinc feed supplementation of piglets increases the proportion of multi-resistant Escherichia coli in vivo. International Journal of Medical Microbiology, 303, 396–403. https://doi.org/10.1016/j.ijmm.2013.06.004
Beier, R. C., Anderson, P. N., Hume, M. E., Poole, T. L., Duke, S. E., Crippen, T. L., Sheffield, C. L., Caldwell, D. J., Byrd, J. A., Anderson, R. C., & Nisbet, D. J. (2011). Characterization of salmonella enterica isolates from turkeys in commercial processing plants for resistance to antibiotics, disinfectants, and a growth promoter. Foodborne Pathogens and Disease, 8(5), 593–600. https://doi.org/10.1089/fpd.2010.0702
Beier, R. C., Byrd, J. A., Andrews, K., Caldwell, D., Crippen, T. L., Anderson, R. C., & Nisbet, D. J. (2021). Disinfectant and antimicrobial susceptibility studies of the foodborne pathogen Campylobacter jejuni isolated from the litter of broiler chicken houses. Poultry Science, 100(2), 1024-1033. https://doi.org/10.1016/j.psj.2020.10.045
Beier, R. C., Harvey, R. B., Hernandez, C. A., Andrews, K., Droleskey, R. E., Hume, M. E., Davidson, M. K., Bodeis-Jones, S., Young, S., Anderson, R. C., & Nisbet, D. J. (2019). Disinfectant and antimicrobial susceptibility profiles of Campylobacter coli isolated in 1998 to 1999 and 2015 from swine and commercial pork chops. Journal of Food Science, 84(6), 1501-1512. https://doi.org/10.1111/1750-3841.14622
Beier, R. C., Poole, T. L., Brichta-Harhay, D. M., Anderson, R. C., Bischoff, K. M., Hernandez, C. A., Bono, J. L., Arthur, T. M., Nagaraja, T. G., Crippen, T. L., Sheffield, C. L., & Nisbet, D. J. (2013). Disinfectant and antibiotic susceptibility profiles of Escherichia coli O157: H7 strains from cattle carcasses, feces, and hides and ground beef from the United States. Journal of Food Protection, 76(1), 6-17. https://doi.org/10.4315/0362-028X.JFP-12-253
Bell, N. J., Potterton, S., Blowey, R., Whay, H. R., & Huxley, J. N. (2014). Disinfectant footbathing agents for the control of bovine digital dermatitis in dairy cattle. Livestock, 19(1), 6-13. https://doi.org/10.12968/live.2014.19.1.6
Bengtsson-Palme, J. (2017). Antibiotic resistance in the food supply chain: where can sequencing and metagenomics aid risk assessment? Current Opinion in Food Science, 14, 66-71. https://doi.org/10.1016/j.cofs.2017.01.010
Bennett, P. M. (2008). Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. British Journal of Pharmacology, 153(S1) S347-S357. https://doi.org/10.1038/sj.bjp.0707607
Bennett, P. M., Livesey, C. T., Nathwani, D., Reeves, D. S., Saunders, J. R., & Wise, R. (2004). An assessment of the risks associated with the use of antibiotic resistance genes in genetically modified plants: report of the Working Party of the British Society for Antimicrobial Chemotherapy. Journal of Antimicrobial Chemotherapy, 53(3), 418-431. https://doi.org/10.1093/jac/dkh087
Bloomfield, S. F. (2002). Significance of biocide usage and antimicrobial resistance in domiciliary environments. Journal of Applied Microbiology, 92, 144S-157S. https://doi.org/10.1046/j.1365-2672.92.5s1.15.x
Bridier, A., Le Grandois, P., Moreau, M. H., Prénom, C., Le Roux, A., Feurer, C., & Soumet, C. (2019). Impact of cleaning and disinfection procedures on microbial ecology and Salmonella antimicrobial resistance in a pig slaughterhouse. Scientific Reports, 9(1), 1-13. https://doi.org/10.1038/s41598-019-49464-8
Burridge, L., Weis, J. S., Cabello, F., Pizarro, J., & Bostick, K. (2010). Chemical use in salmon aquaculture: a review of current practices and possible environmental effects. Aquaculture, 306(1-4), 7-23. https://doi.org/10.1016/j.aquaculture.2010.05.020
Buta, M., Korzeniewska, E., Harnisz, M., Hubeny, J., Zieliński, W., Rolbiecki, D.,
Bajkacz, S., Felis, E., & Kokoszka, K. (2021). Microbial and chemical pollutants on the manure-crops pathway in the perspective of “One Health” holistic approach. Science of the Total Environment, 785, 147411. https://doi.org/10.1016/j.scitotenv.2021.147411
Cabello, F. C., Godfrey, H. P., Buschmann, A. H., & Dölz, H. J. (2016). Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. The Lancet Infectious Diseases, 16(7), e127-e133. https://doi.org/10.1016/S1473-3099(16)00100-6
Capita, R., & Alonso-Calleja, C. (2013). Antibiotic-resistant bacteria: a challenge for the food industry. Critical Reviews in Food Science and Nutrition, 53(1), 11-48. https://doi.org/10.1080/10408398.2010.519837
Capita, R., Álvarez-Fernández, E., Fernández-Buelta, E., Manteca, J., & Alonso Calleja, C. (2013). Decontamination treatments can increase the prevalence of resistance to antibiotics of Escherichia coli naturally present on poultry. Food Microbiology, 34(1), 112-117. http://dx.doi.org/10.1016/j.fm.2012.11.011
Carattoli, A. (2001). Importance of integrons in the diffusion of resistance. Veterinary Research, 32(3-4), 243-259. https://doi.org/10.1051/vetres:2001122
Caruana, J. C., & Walper, S. A. (2020). Bacterial membrane vesicles as mediators of microbe–microbe and microbe–host community interactions. Frontiers in Microbiology, 11, 432. https://doi.org/10.3389/fmicb.2020.00432
Cavaco, L. M., Hasman, H., & Aarestrup, F. M. (2011). Zinc resistance of Staphylococcus aureus of animal origin is strongly associated with methicillin resistance. Veterinary Microbiology, 150(3-4), 344-348. https://doi.org/10.1016/j.vetmic.2011.02.014
Cavaco, L. M., Hasman, H., Stegger, M., Andersen, P. S., Skov, R., Fluit, A. C., Ito, T., & Aarestrup, F. M. (2010). Cloning and occurrence of czrC, a gene conferring cadmium and zinc resistance in methicillin-resistant Staphylococcus aureus CC398 isolates. Antimicrobial Agents and Chemotherapy, 54(9), 3605-3608. https://doi.org/10.1128/AAC.00058-10
Cheng, G., Ning, J., Ahmed, S., Huang, J., Ullah, R., An, B., Hao, H., Dai, M., Huang, L., Wang, X., & Yuan, Z. (2019). Selection and dissemination of antimicrobial resistance in Agri-food production. Antimicrobial Resistance & Infection Control, 8(1), 1-13. https://doi.org/10.1186/s13756-019-0623-2
Chenia, H. Y., & Jacobs, A. (2017). Antimicrobial resistance, heavy metal resistance and integron content in bacteria isolated from a South African tilapia aquaculture system. Diseases of Aquatic Organisms, 126(3), 199-209. https://doi.org/10.3354/dao03173
Chuanchuen, R., Pathanasophon, P., Khemtong, S., Wannaprasat, W., & Padungtod, P. (2008). Susceptibilities to antimicrobials and disinfectants in Salmonella isolates obtained from poultry and swine in Thailand. Journal of Veterinary Medical Science, 70(6), 595-601. https://doi.org/10.1292/jvms.70.595
Ciesinski, L., Guenther, S., Pieper, R., Kalisch, M., Bednorz, C., & Wieler, L. H. (2018). High dietary zinc feeding promotes persistence of multi-resistant E. coli in the swine gut. PLoS One, 13(1), e0191660. https://doi.org/10.1371/journal.pone.0191660
Ciric, L., Mullany, P., & Roberts, A. P. (2011). Antibiotic and antiseptic resistance genes are linked on a novel mobile genetic element: Tn 6087. Journal of Antimicrobial Chemotherapy, 66(10), 2235-2239. https://doi.org/10.1093/jac/dkr311
Cogliani, C., Goossens, H., & Greko, C. (2011). Restricting antimicrobial use in food animals: Lessons from Europe. Microbe, 6(6), 274–279. https://doi.org/10.1128/microbe.6.274.1
Colavecchio, A., Cadieux, B., Lo, A., & Goodridge, L. D. (2017). Bacteriophages contribute to the spread of antibiotic resistance genes among foodborne pathogens of the Enterobacteriaceae family–a review. Frontiers in Microbiology, 8, 1108. https://doi.org/10.3389/fmicb.2017.01108
Cufaoglu, G., Cengiz, G., Acar, B., Yesilkaya, B., Ayaz, N. D., Levent, G., & Goncuoglu, M. (2022). Antibiotic, heavy metal, and disinfectant resistance in chicken, cattle, and sheep origin E. coli and whole‐genome sequencing analysis of a multidrug‐resistant E. coli O100:H25 strain. Journal of Food Safety, 42(5), e12995. https://doi.org/10.1111/jfs.12995
Davies, R., & Wales, A. (2019). Antimicrobial resistance on farms: a review including biosecurity and the potential role of disinfectants in resistance selection. Comprehensive Reviews in Food Science and Food Safety, 18(3), 753-774. https://doi.org/10.1111/1541-4337.12438
de Souza, E. L. (2016). The effects of sublethal doses of essential oils and their constituents on antimicrobial susceptibility and antibiotic resistance among food-related bacteria: A review. Trends in Food Science & Technology, 56, 1-12. http://dx.doi.org/10.1016/j.tifs.2016.07.012
Dębski, B. (2016). Supplementation of pigs diet with zinc and copper as alternative to conventional antimicrobials. Polish Journal of Veterinary Sciences, 19(4). https://doi.org/10.1515/pjvs-2016-0113
Denyer, S. P., & Maillard, J. Y. (2002). Cellular impermeability and uptake of biocides and antibiotics in Gram‐negative bacteria. Journal of Applied Microbiology, 92(s1), 35S-45S. https://doi.org/10.1046/j.1365-2672.92.5s1.19.x
Diard, M., Bakkeren, E., Cornuault, J. K., Moor, K., Hausmann, A., Sellin, M. E., Loverdo, C., Aertsen, A., Ackermann, M, de Papepe, M., Slack, E., & Hardt, W. D. (2017). Inflammation boosts bacteriophage transfer between Salmonella spp. Science 355, 1211–1215. https://doi.org/10.1126/science.aaf8451
Donaghy, J. A., Jagadeesan, B., Goodburn, K., Grunwald, L., Jensen, O. N., Jespers, A. D., Kanagachandran, K., Lafforgue, H., Seefelder, W., & Quentin, M. C. (2019). Relationship of sanitizers, disinfectants, and cleaning agents with antimicrobial resistance. Journal of Food Protection, 82(5), 889-902. https://doi.org/10.4315/0362-028X.JFP-18-373
Dong, Z., Wang, J., Wang, L., Zhu, L., Wang, J., Zhao, X., & Kim, Y. M. (2022). Distribution of quinolone and macrolide resistance genes and their co-occurrence with heavy metal resistance genes in vegetable soils with long-term application of manure. Environmental Geochemistry and Health, 44(10), 3343-3358. https://doi.org/10.1007/s10653-021-01102-x
Duan, M., Gu, J., Wang, X., Li, Y., Zhang, R., Hu, T., & Zhou, B. (2019). Factors that affect the occurrence and distribution of antibiotic resistance genes in soils from livestock and poultry farms. Ecotoxicology and Environmental Safety, 180, 114-122. https://doi.org/10.1016/j.ecoenv.2019.05.005
Dweba, C. C., Zishiri, O. T., & El Zowalaty, M. E. (2018). Methicillin-resistant Staphylococcus aureus: livestock-associated, antimicrobial, and heavy metal resistance. Infection and Drug Resistance, 11, 2497. https://doi.org/10.2147/IDR.S175967
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards) (2008). Assessment of the possible effect of the four antimicrobial treatment substances on the emergence of antimicrobial resistance. EFSA Journal, 659, https://doi.org/10.2903/j.efsa.2008.659
EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards) (2021). Scientific Opinion on the role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA Journal,19(6):6651, 188 pp. https://doi.org/10.2903/j.efsa.2021.6651
EFSA FEEDAP Panel (EFSA Panel on Additives and Products or Substances used in Animal Feed) (2014). Scientific opinion on the potential reduction of the currently authorised maximum zinc content in complete feed. EFSA Journal,12(5):3668, 77 pp. https://doi.org/10.2903/j.efsa.2014.3668
EFSA FEEDAP Panel (EFSA Panel on Additives and Products or Substances used in Animal Feed) (2016). Scientific opinion on the revision of the currently authorised maximum copper content in complete feed. EFSA Journal, 14(8):4563, 100 pp. https://doi.org/10.2903/j.efsa.2016.4563
Ejileugha, C. (2022). Biochar can mitigate co-selection and control antibiotic resistant genes (ARGs) in compost and soil. Heliyon, e09543. https://doi.org/10.1016/j.heliyon.2022.e09543
El Behiry, A., Schlenker, G., Szabo, I., & Roesler, U. (2012). In vitro susceptibility of Staphylococcus aureus strains isolated from cows with subclinical mastitis to different antimicrobial agents. Journal of Veterinary Science, 13(2), 153-161. https://doi.org/10.4142/jvs.2012.13.2.153
Elbehiry, A., Al‐Dubaib, M., Marzouk, E., & Moussa, I. (2019). Antibacterial effects and resistance induction of silver and gold nanoparticles against Staphylococcus aureus‐induced mastitis and the potential toxicity in rats. MicrobiologyOpen, 8(4), e00698. https://doi.org/10.1002/mbo3.698
EMA & EFSA (European Medicines Agency and European Food Safety Authority) (2017). EMA and EFSA Joint Scientific Opinion on measures to reduce the need to use antimicrobial agents in animal husbandry in the European Union, and the resulting impacts on food safety (RONAFA). [EMA/CVMP/570771/2015]. EFSA Journal, 15(1):4666, 245 pp. https://doi.org/10.2903/j.efsa. 2017.4666
Eom, H. S., Back, S. H., Lee, H. H., Lee, G. Y., & Yang, S. J. (2019). Prevalence and characteristics of livestock-associated methicillin-susceptible Staphylococcus aureus in the pork production chain in Korea. Journal of Veterinary Science, 20(6), e69. https://doi.org/10.4142/jvs.2019.20.e69
Ezugworie, F. N., Igbokwe, V. C., & Onwosi, C. O. (2021). Proliferation of antibiotic-resistant microorganisms and associated genes during composting: An overview of the potential impacts on public health, management and future. Science of The Total Environment, 784, 147191. https://doi.org/10.1016/j.scitotenv.2021.147191
Fadli, M., Chevalier, J., Hassani, L., Mezrioui, N. E., & Pages, J. M. (2014). Natural extracts stimulate membrane-associated mechanisms of resistance in Gram-negative bacteria. Letters in Applied Microbiology, 58, 472–477. https://doi.org/10.1111/lam.12216
Fang, L., Li, X., Li, L., Li, S., Liao, X., Sun, J., & Liu, Y. (2016). Co-spread of metal and antibiotic resistance within ST3-IncHI2 plasmids from E. coli isolates of food-producing animals. Scientific Reports, 6(1), 1-8. https://doi.org/10.1038/srep25312.
FAO/WHO (2019). Joint FAO/WHO Expert Meeting in Collaboration with OIE on Foodborne Antimicrobial Resistance: Role of the Environment. Crops and Biocides. Microbiological Risk Assessment Series no. 34, Rome. (Last accessed: February 2023)
Ferreira, J. C., Penha Filho, R. A. C., Andrade, L. N., & Darini, A. L. C. (2019). Evaluation of heavy metal tolerance genes in plasmids harbored in multidrug-resistant Salmonella enterica and Escherichia coli isolated from poultry in Brazil. Diagnostic Microbiology and Infectious Disease, 94(3), 314-315. https://doi.org/10.1016/j.diagmicrobio.2019.01.019
Feßler, A. T., Zhao, Q., Schoenfelder, S., Kadlec, K., Michael, G. B., Wang, Y., Ziebuhr, W., Shen, J., & Schwarz, S. (2017). Complete sequence of a plasmid from a bovine methicillin-resistant Staphylococcus aureus harbouring a novel ica-like gene cluster in addition to antimicrobial and heavy metal resistance genes. Veterinary Microbiology, 200, 95-100. https://doi.org/10.1016/j.vetmic.2016.07.010
Food Standards Agency (2016). Chief Scientific Adviser’s Science Report, Issue four: Antimicrobial resistance in the food supply chain. Food Standards Agency. (Last accessed: December 2022)
Galetti, R., Penha Filho, R. A. C., Ferreira, J. C., Varani, A. M., & Darini, A. L. C. (2019). Antibiotic resistance and heavy metal tolerance plasmids: The antimicrobial bulletproof properties of Escherichia fergusonii isolated from poultry. Infection and Drug Resistance, 12, 1029-1033. https://doi.org/10.2147/IDR.S196411
Galetti, R., Penha Filho, R. A. C., Ferreira, J. C., Varani, A. M., Sazinas, P., Jelsbak, L., & Darini, A. L. C. (2021). The plasmidome of multidrug-resistant emergent Salmonella serovars isolated from poultry. Infection, Genetics and Evolution, 89, 104716. https://doi.org/10.1016/j.meegid.2021.104716
Gantzhorn, M. R., Pedersen,K., Olsen, J. E., & Thomsen, L. E. (2014). Biocide and antibiotic susceptibility of Salmonella isolates obtained before and after cleaning at six Danish pig slaughterhouses. International Journal of Food Microbiology, 181, 53–59. https://doi.org/10.1016/j.ijfoodmicro.2014.04.021
Geueke, B. (2014). Dossier – Biocides and food contact materials. Food Packaging Forum. (Last accessed: April 2023)
Ghazisaeedi, F., Ciesinski, L., Bednorz, C., Johanns, V., Pieper, L., Tedin, K., Wieler, L. H., & Günther, S. (2020). Phenotypic zinc resistance does not correlate with antimicrobial multi-resistance in fecal E. coli isolates of piglets. Gut pathogens, 12(1), 1-10. https://doi.org/10.1186/s13099-019-0342-5
Giacometti, F., Shirzad-Aski, H., & Ferreira, S. (2021). Antimicrobials and food-related stresses as selective factors for antibiotic resistance along the farm to fork continuum. Antibiotics, 10(6), 671. https://doi.org/10.3390/antibiotics10060671
Gomez-Sanz, E., Kadlec, K., Fessler, A. T., Zarazaga, M., Torres, C., & Schwarz, S. (2013). Novel erm(T)-carrying multiresistance plasmids from porcine and human isolates of methicillin-resistant Staphylococcus aureus ST398 that also harbor cadmium and copper resistance determinants. Antimicrobial Agents and Chemotherapy, 57(7), 3275–3282. https://doi.org/10.1128/AAC.00171-13
Guardiola, F. A., Cuesta, A., Meseguer, J., & Esteban, M. A. (2012). Risks of using antifouling biocides in aquaculture. International Journal of Molecular Sciences, 13(2), 1541-1560. https://doi.org/10.3390/ijms13021541
Gullberg, E., Albrecht, L. M., Karlsson, C., Sandegren, L., & Andersson, D. I. (2014). Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. MBio, 5(5), e01918-14. https://doi.org/10.1128/mBio.01918-14
Guo, T., Lou, C. L., Zhai, W. W., Tang, X. J., Hashmi, M. Z., Murtaza, R., Li, Y., Liu, X. M., & Xu, J. M. (2018). Increased occurrence of heavy metals, antibiotics and resistance genes in surface soil after long-term application of manure. The Science of the Total Environment, 635, 995–1003. https://doi.org/10.1016/j.scitotenv.2018.04.194
Haley, B. J., Kim, S. J., Biswas, D., Hovingh, E., & Van Kessel, J. A. S. (2022). Virulome and genome analyses identify associations between antimicrobial resistance genes and virulence factors in highly drug-resistant Escherichia coli isolated from veal calves. PLOS ONE, 17(3), e0265445. https://doi.org/10.1371/journal.pone.0265445
Hall, J. P., Brockhurst, M. A., & Harrison, E. (2017). Sampling the mobile gene pool: innovation via horizontal gene transfer in bacteria. Philosophical Transactions of the Royal Society B: Biological Sciences, 372(1735), 20160424. https://doi.org/10.1098/rstb.2016.0424
Hall, J. P., Wright, R. C., Guymer, D., Harrison, E., & Brockhurst, M. A. (2020). Extremely fast amelioration of plasmid fitness costs by multiple functionally diverse pathways. Microbiology, 166(1), 56-62. https://doi.org/10.1099/mic.0.000862
Hall, J. P., Wright, R. C., Harrison, E., Muddiman, K. J., Wood, A. J., Paterson, S., & Brockhurst, M. A. (2021). Plasmid fitness costs are caused by specific genetic conflicts enabling resolution by compensatory mutation. PLoS biology, 19(10), e3001225. https://doi.org/10.1371/journal.pbio.3001225
Hasman, H., & Aarestrup, F. M. (2002). tcrB a gene conferring transferable copper resistance in Enterococcus faecium: Occurrence, transferability, and linkage to macrolide and glycopeptide resistance. Antimicrobial Agents and Chemotherapy, 46, 1410–1416. https://doi.org/10.1128/AAC.46.5.1410-1416.2002
Hasman, H., Kempf, I., Chidaine, B., Cariolet, R., Ersbøll, A. K., Houe, H., Hansen, H. C. B., & Aarestrup, F. M. (2006). Copper resistance in Enterococcus faecium, mediated by the tcrB gene, is selected by supplementation of pig feed with copper sulfate. Applied and Environmental Microbiology, 72(9), 5784-5789. https://doi.org/10.1128/AEM.02979-05
Hau, S. J., Frana, T., Sun, J., Davies, P. R., & Nicholson, T. L. (2017). Zinc resistance within swine-associated methicillin-resistant Staphylococcus aureus isolates in the United States is associated with multilocus sequence type lineage. Applied and Environmental Microbiology, 83, 1–9. https://doi.org/10.1128/AEM.00756-17
Hegstad, K., Langsrud, S., Lunestad, B. T., Scheie, A. A., Sunde, M., & Yazdankhah, S. P. (2010). Does the wide use of quaternary ammonium compounds enhance the selection and spread of antimicrobial resistance and thus threaten our health? Microbial Drug Resistance, 16(2), 91-104. https://doi.org/10.1089/mdr.2009.0120
Hejna, M., Gottardo, D., Baldi, A., Dell’Orto, V., Cheli, F., Zaninelli, M., & Rossi, L. (2018). Review: Nutritional ecology of heavy metals. Animal, 12(10), 2156-2170. https://doi.org/10.1017/S175173111700355X
Hölzel, C. S., Müller, C., Harms, K. S., Mikolajewski, S., Schäfer, S., Schwaiger, K., & Bauer, J. (2012). Heavy metals in liquid pig manure in light of bacterial antimicrobial resistance. Environmental Research, 113, 21-27. https://doi.org/10.1016/j.envres.2012.01.002
Hölzel, C. S., Tetens, J. L., & Schwaiger, K. (2018). Unraveling the role of vegetables in spreading antimicrobial-resistant bacteria: a need for quantitative risk assessment. Foodborne Pathogens and Disease. 15(11), 671-688. https://doi.org/10.1089/fpd.2018.2501
Hu, Y., Jiang, L., Zhang, T., Jin, L., Han, Q., Zhang, D., Lin, K., & Cui, C. (2018). Occurrence and removal of sulfonamide antibiotics and antibiotic resistance genes in conventional and advanced drinking water treatment processes. Journal of Hazardous Materials, 360, 364-372. https://doi.org/10.1016/j.jhazmat.2018.08.012
Huddleston, J. R. (2014). Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infection and Drug Resistance, 7, 167. https://doi.org/10.2147/IDR.S48820
Jacob, M. E., Fox, J. T., Nagaraja, T. G., Drouillard, J. S., Amachawadi, R. G., & Narayanan, S. K. (2010). Effects of feeding elevated concentrations of copper and zinc on the antimicrobial susceptibilities of fecal bacteria in feedlot cattle. Foodborne Pathogens and Disease, 7(6), 643-648. https://doi.org/10.1089/fpd.2009.0401
James, C., Dixon, R., Talbot, L., James, S. J., Williams, N., & Onarinde, B. A. (2021). Assessing the impact of heat treatment of food on antimicrobial resistance genes and their potential uptake by other bacteria - A critical review. Antibiotics, 10(12) p1440. https://doi.org/10.3390/antibiotics10121440
Jebri, S., Rahmani, F., & Hmaied, F. (2021). Bacteriophages as antibiotic resistance genes carriers in agro‐food systems. Journal of Applied Microbiology, 130(3), 688-698. https://doi.org/10.1111/jam.14851
Jensen, J., Kyvsgaard, N. C., Battisti, A., & Baptiste, K. E. (2018). Environmental and public health related risk of veterinary zinc in pig production-using Denmark as an example. Environment International, 114, 181-190. https://doi.org/10.1016/j.envint.2018.02.007
Ji, X., Shen, Q., Liu, F., Ma, J., Xu, G., Wang, Y., & Wu, M. (2012). Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai; China. Journal of Hazardous Materials, 235, 178-185. https://doi.org/10.1016/j.jhazmat.2012.07.040
Jones, I. A., & Joshi, L. T. (2021). Biocide use in the antimicrobial era: A review. Molecules, 26(8), 2276. https://doi.org/10.3390/molecules26082276
Jutkina, J., Marathe, N. P., Flach, C.-F., & Larsson, D. G. J. (2018). Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations. Science of the Total Environment, 616–617, 172–178. https://doi.org/10.1016/j.scitotenv.2017.10.312
Kampf, G. (2018). Biocidal agents used for disinfection can enhance antibiotic resistance in gram-negative species. Antibiotics, 7(4), 110. https://doi.org/10.3390/antibiotics7040110
Kampf, G. (2019). Antibiotic resistance can be enhanced in Gram-positive species by some biocidal agents used for disinfection. Antibiotics, 8(1), 13. https://doi.org/10.3390/antibiotics8010013
Klümper, U., Recker, M., Zhang, L., Yin, X., Zhang, T., Buckling, A., & Gaze, W. H. (2019). Selection for antimicrobial resistance is reduced when embedded in a natural microbial community. The ISME journal, 13(12), 2927-2937. https://doi.org/10.1038/s41396-019-0483-z
Kotb, S., & Sayed, M. (2015). Sensitivity of methicillin-resistance and methicillin-susceptible Staphylococcus aureus strains to some different disinfectants. International Journal of Livestock Research, 5(8), 45–58. https://doi.org/10.5455/ijlr.20150822031340
Kulkarni, H. M., Nagaraj, R., & Jagannadham, M. V. (2015). Protective role of E. coli outer membrane vesicles against antibiotics. Microbiological Research, 181, 1-7. https://doi.org/10.1016/j.micres.2015.07.008
Langsrud, S., Møretrø, T., & Sundheim, G. (2003). Characterization of Serratia marcescens surviving in disinfecting footbaths. Journal of Applied Microbiology, 95(1), 186-195. https://doi.org/10.1046/j.1365-2672.2003.01968.x
Le Devendec, L., Jouy, E., & Kempf, I. (2018). Evaluation of resistance gene transfer from heat-treated Escherichia coli. International Journal of Food Microbiology, 270, 39-43. https://doi.org/10.1016/j.ijfoodmicro.2018.02.019
Lemire, J. A., Harrison, J. J., & Turner, R. J. (2013). Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nature Reviews Microbiology, 11(6), 371-384. https://doi.org/10.1038/nrmicro3028
Li, C., Quan, Q., Gan, Y., Dong, J., Fang, J., Wang, L., & Liu, J. (2020). Effects of heavy metals on microbial communities in sediments and establishment of bioindicators based on microbial taxa and function for environmental monitoring and management. Science of the Total Environment, 749, 141555. https://doi.org/10.1016/j.scitotenv.2020.141555
Li, N., Chen, J., Liu, C., Yang, J., Zhu, C., & Li, H. (2022c). Cu and Zn exert a greater influence on antibiotic resistance and its transfer than doxycycline in agricultural soils. Journal of Hazardous Materials, 423, 127042. https://doi.org/10.1016/j.jhazmat.2021.127042
Li, N., Li, H., Zhu, C., Liu, C., Su, G., & Chen, J. (2022a). Controlling AMR in the pig industry: Is it enough to restrict heavy metals?. International Journal of Environmental Research and Public Health, 19(18), 11265. https://doi.org/10.3390/ijerph191811265
Li, Y., Liu, B., Zhang, X., Gao, M., & Wang, J. (2015). Effects of Cu exposure on enzyme activities and selection for microbial tolerances during swine-manure composting. Journal of Hazardous Materials, 283, 512-518. https://doi.org/10.1016/j.jhazmat.2014.09.061
Li, Z., Junaid, M., Chen, G., & Wang, J. (2022b). Interactions and associated resistance development mechanisms between microplastics, antibiotics and heavy metals in the aquaculture environment. Reviews in Aquaculture, 14(2), 1028-1045. https://doi.org/10.1111/raq.12639
Likotrafiti, E., Oniciuc, E. A., Prieto, M., Santos, J. A., López, S., & Alvarez‐Ordóñez, A. (2018). Risk assessment of antimicrobial resistance along the food chain through culture‐independent methodologies. EFSA Journal, 16(S1), e160811. https://doi.org/10.2903/j.efsa.2018.e160811
Liu, B., Yu, K., Ahmed, I., Gin, K., Xi, B., Wei, Z., He, Y., & Zhang, B. (2021a). Key factors driving the fate of antibiotic resistance genes and controlling strategies during aerobic composting of animal manure: A review. Science of the Total Environment, 791, 148372. https://doi.org/10.1016/j.scitotenv.2021.148372
Liu, C., Li, G., Qin, X., Xu, Y., Wang, J., Wu, G., Feng, H., Ye, J., Zhu, C., Li, X., & Zheng, X. (2022). Profiles of antibiotic-and heavy metal-related resistance genes in animal manure revealed using a metagenomic analysis. Ecotoxicology and Environmental Safety, 239, 113655. https://doi.org/10.1016/j.ecoenv.2022.113655
Liu, C., Li, X., Zheng, S., Kai, Z., Jin, T., Shi, R., Huang, H., & Zheng, X. (2021b). Effects of wastewater treatment and manure application on the dissemination of antimicrobial resistance around swine feedlots. Journal of Cleaner Production, 280, 123794. https://doi.org/10.1016/j.jclepro.2020.123794
Liwa, A., & Jaka, H. M. (2015). Antimicrobial resistance: Mechanisms of action of antimicrobial agents. In: The Battle Against Microbial Pathogens: Basic Science, Technological Advances and Educational Programs, A. Méndez-Vilas (Ed), Vol. 1, 876-885. (Last accessed: April 2023)
Lorenz, M. G., & Wackernagel, W. (1994). Bacterial gene transfer by natural genetic transformation in the environment. Microbiological Reviews, 58(3), 563-602. https://doi.org/10.1128/mr.58.3.563-602.1994.
Maertens, H., De Reu, K., Meyer, E., Van Coillie, E., & Dewulf, J. (2019). Limited association between disinfectant use and either antibiotic or disinfectant susceptibility of Escherichia coli in both poultry and pig husbandry. BMC Veterinary Research, 15(1), 1-12. https://doi.org/10.1186/s12917-019-2044-0
Maertens, H., De Reu, K., Meyer, E., Van Weyenberg, S., Dewulf, J., & Van Coillie, E. (2019). Exposure of ciprofloxacin-resistant Escherichia coli broiler isolates to subinhibitory concentrations of a quaternary ammonium compound does not increase antibiotic resistance gene transfer. Poultry Science, 98(7), 2972-2976. http://dx.doi.org/10.3382/ps/pez185
Maertens, H., Van Coillie, E., Millet, S., Van Weyenberg, S., Sleeckx, N., Meyer, E., Zoons, J., Dewulf, J., & De Reu, K. (2020). Repeated disinfectant use in broiler houses and pig nursery units does not affect disinfectant and antibiotic susceptibility in Escherichia coli field isolates. BMC Veterinary Research, 16, 1-11. https://doi.org/10.1186/s12917-020-02342-2
Maillard, J. Y. (2018). Resistance of bacteria to biocides. Microbiology Spectrum, 6(2), ARBA-0006-2017. https://doi.org/10.1128 /microbiolspec.ARBA-0006-2017
Maillard, J. Y. (2020). Bacterial resistance to biocides. In: McDonnell, G. and Hansen, J. (Eds) Blocks’ Disinfection, Sterilization and Preservation, 6th edn. Philadelphia: Wolters Kluwer. (Last accessed: April 2023)
Maillard, J. Y., & Hartemann, P. (2013). Silver as an antimicrobial: facts and gaps in knowledge. Critical Reviews in Microbiology, 39(4), 373-383. https://doi.org/10.3109/1040841X.2012.713323
Mariotti, M., Lombardini, G., Rizzo, S., Scarafile, D., Modesto, M., Truzzi, E., Benvenuti, S., Elmi, A., Bertocchi, M., Fiorentini, L., Gambi, L., Scozzoli, M., & Mattarelli, P. (2022). Potential applications of essential oils for environmental sanitization and antimicrobial treatment of intensive livestock infections. Microorganisms, 10(4), 822. https://doi.org/10.3390/microorganisms10040822
Mavri, A., Kurinčič, M., & Možina, S. S. (2012). The prevalence of antibiotic and biocide resistance among Campylobacter coli and Campylobacter jejuni from different sources. Food Technology & Biotechnology, 50(3), 371-376. (Last accessed: February 2023)
Mazhar, S. H., Li, X., Rashid, A., Su, J., Xu, J., Brejnrod, A. D., Su, J., Wu, Y., Zhu, Y., Zhou, S., Feng, R., & Rensing, C. (2021). Co-selection of antibiotic resistance genes, and mobile genetic elements in the presence of heavy metals in poultry farm environments. Science of the Total Environment, 755, 142702. https://doi.org/10.1016/j.scitotenv.2020.142702
McDonnell, G., & Russell, A. D. (1999). Antiseptics and disinfectants: activity, action, and resistance. Clinical Microbiology Reviews, 12(1), 147-179. https://doi.org/10.1128/CMR.12.1.147
McMahon, M. A. S., Blair, I. S., Moore, J. E., & McDowell, D. A. (2007b). Habituation to sub-lethal concentrations of tea tree oil (Melaleuca alternifolia) is associated with reduced susceptibility to antibiotics in human pathogens. Journal of Antimicrobial Chemotherapy, 59, 125–127. https://doi.org/10.1093/jac/dkl443
McMahon, M. A. S., Xu, J., Moore, J. E., Blair, I. S., & McDowell, D. A. (2007a). Environmental stress and antibiotic resistance in food-related pathogens. Applied Environmental Microbiology, 73, 211–217. https://doi.org/10.1128/AEM.00578-06
Medardus, J. J., Molla, B. Z., Nicol, M., Morrow, W. M., Rajala-Schultz, P. J., Kazwala, R., & Gebreyes, W. A. (2014). In-feed use of heavy metal micronutrients in US swine production systems and its role in persistence of multidrug-resistant salmonellae. Applied and Environmental Microbiology, 80(7), 2317-2325. https://doi.org/10.1128/AEM.04283-13
Molina-Gonzalez, D., Alonso-Calleja, C., Alonso-Hernando, A., & Capita, R. (2014). Effect of sub-lethal concentrations of biocides on the susceptibility to antibiotics of multi-drug resistant Salmonella enterica strains. Food Control, 40, 329-334. http://dx.doi.org/10.1016/j.foodcont.2013.11.046
Montagnin, C., Cawthraw, S., Ring, I., Ostanello, F., Smith, R. P., Davies, R., & Martelli, F. (2022). Efficacy of five disinfectant products commonly used in pig herds against a panel of bacteria sensitive and resistant to selected antimicrobials. Animals, 12(20), 2780. https://doi.org/10.3390/ani12202780
Mozaheb, N., & Mingeot-Leclercq, M. P. (2020). Membrane vesicle production as a bacterial defense against stress. Frontiers in Microbiology, 11, 600221. https://doi.org/10.3389/fmicb.2020.600221
Mulder, I., Siemens, J., Sentek, V., Amelung, W., Smalla, K., & Jechalke, S. (2018). Quaternary ammonium compounds in soil: implications for antibiotic resistance development. Reviews in Environmental Science and Bio/Technology, 17(1), 159-185. https://doi.org/10.1007/s11157-017-9457-7
Munita, J. M., & Arias, C. A. (2016). Mechanisms of antibiotic resistance. Microbiology Spectrum, 4(2), 4-2. https://doi.org/10.1128/microbiolspec.VMBF-0016-2015
Murray, S. A., Amachawadi, R. G., Norman, K. N., Lawhon, S. D., Nagaraja, T. G., Drouillard, J. S., & Scott, H. M. (2021). Effects of zinc and menthol-based diets on co-selection of antibiotic resistance among E. coli and Enterococcus spp. in beef cattle. Animals, 11(2), 259. https://doi.org/10.3390/ani11020259
Nhung, N. T., Thuy, C. T., Trung, N. V., Campbell, J., Baker, S., Thwaites, G., Hoa, N. T., & Carrique-Mas, J. (2015). Induction of antimicrobial resistance in Escherichia coli and non-typhoidal Salmonella strains after adaptation to disinfectant commonly used on farms in Vietnam. Antibiotics, 4(4), 480-494. https://doi.org/10.3390/antibiotics4040480
Nicholson, F. A., Chambers, B. J., Williams, J. R., & Unwin, R. J. (1999). Heavy metal contents of livestock feeds and animal manures in England and Wales. Bioresource Technology, 70(1), 23-31. https://doi.org/10.1016/S0960-8524(99)00017-6
Nicholson, F. A., Smith, S. R., Alloway, B. J., Carlton-Smith, C., & Chambers, B. J. (2003). An inventory of heavy metals inputs to agricultural soils in England and Wales. Science of the Total Environment, 311(1-3), 205-219. https://doi.org/10.1016/S0048-9697(03)00139-6
Nicholson, F. A., Smith, S. R., Alloway, B. J., Carlton‐Smith, C., & Chambers, B. J. (2006). Quantifying heavy metal inputs to agricultural soils in England and Wales. Water and Environment Journal, 20(2), 87-95. https://doi.org/10.1111/j.1747-6593.2006.00026.x
O’Neill, J. (2016). Tackling drug-resistant infections globally: Final report and recommendations. (Last accessed: February 2023)
Ortega Morente, E. O., Fernández-Fuentes, M. A., Burgos, M. J. G., Abriouel, H., Pulido, R. P., & Gálvez, A. (2013). Biocide tolerance in bacteria. International Journal of Food Microbiology, 162(1), 13-25. http://dx.doi.org/10.1016/j.ijfoodmicro.2012.12.028
Pal, C., Bengtsson-Palme, J., Kristiansson, E., & Larsson, D. G. (2015). Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics, 16, 964. https://doi.org/10.1186/s12864-015-2153-5
Pearce, H., Messager, M., & Maillard, J. Y. (1999). Effect of biocides commonly used in the hospital environment on the transfer of antibiotic-resistance genes in Staphylococcus aureus. Journal of Hospital Infection, 43(2), 101–107. https://doi.org/10.1053/jhin.1999.0250
Peng, S., Feng, Y., Wang, Y., Guo, X., Chu, H., & Lin, X. (2017). Prevalence of antibiotic resistance genes in soils after continually applied with different manure for 30 years. Journal of Hazardous Materials, 340, 16-25. https://doi.org/10.1016/j.jhazmat.2017.06.059
Peng, S., Zheng, H., Herrero-Fresno, A., Olsen, J. E., Dalsgaard, A., & Ding, Z. (2021). Co-occurrence of antimicrobial and metal resistance genes in pig feces and agricultural fields fertilized with slurry. Science of the Total Environment, 792, 148259. https://doi.org/10.1016/j.scitotenv.2021.148259
Pérez-Rodríguez, F., & Mercanoglu Taban, B. (2019). A state-of-art review on multi-drug resistant pathogens in foods of animal origin: risk factors and mitigation strategies. Frontiers in Microbiology, 10, 2091. https://doi.org/10.3389/fmicb.2019.02091
Petrovska, L., Mather, A. E., AbuOun, M., Branchu, P., Harris, S. R., Connor, T., Hopkins, K. L., Underwood, A., Lettini, A. A.,
Page, A., Bagnall, M., Wain, J., Parkhill, J., Dougan, G., Davies, R., & Kingsley, R. A. (2016). Microevolution of monophasic Salmonella typhimurium during epidemic, United Kingdom, 2005–2010. Emerging Infectious Diseases, 22(4), 617-624. https://doi.org/10.3201/eid2204.150531
Peyrat, M. B., Soumet, C., Maris, P., & Sanders, P. (2008). Phenotypes and genotypes of Campylobacter strains isolated after cleaning and disinfection in poultry slaughterhouses. Veterinary Microbiology, 128(3-4), 313-326. https://doi.org/10.1016/j.vetmic.2007.10.021
Pontin, K. P., Borges, K. A., Furian, T. Q., Carvalho, D., Wilsmann, D. E., Cardoso, H. R. P., Alves, A. K., Chitolina, G. Z., Salle, C. T. P., de Souza Moraes, H. L., & do Nascimento, V. P. (2021). Antimicrobial activity of copper surfaces against biofilm formation by Salmonella Enteritidis and its potential application in the poultry industry. Food Microbiology, 94, 103645. https://doi.org/10.1016/j.fm.2020.103645
Poole, K. (2017). At the nexus of antibiotics and metals: the impact of Cu and Zn on antibiotic activity and resistance. Trends in Microbiology, 25(10), 820-832. https://doi.org/10.1016/j.tim.2017.04.010
Puangseree, J., Jeamsripong, S., Prathan, R., Pungpian, C., & Chuanchuen, R. (2021). Resistance to widely-used disinfectants and heavy metals and cross resistance to antibiotics in Escherichia coli isolated from pigs, pork and pig carcass. Food Control, 124, 107892. https://doi.org/10.1016/j.foodcont.2021.107892
Ragland, D., Schneider, J. L., Amass, S. F., & Hill, M. A. (2006). Alternatives to the use of antimicrobial feed additives in nursery diets: A pilot study. Journal of Swine Health and Production, 14(2), 82-88. (Last accessed: April 2023)
Randall, L. P., Clouting, C. S., Gradel, K. O., Clifton‐Hadley, F. A., Davies, R. D., & Woodward, M. J. (2005). Farm disinfectants select for cyclohexane resistance, a marker of multiple antibiotic resistance, in Escherichia coli. Journal of Applied Microbiology, 98(3), 556-563. https://doi.org/10.1111/j.1365-2672.2004.02488.x
Randall, L. P., Cooles, S. W., Coldham, N. G., Penuela, E. G., Mott, A. C., Woodward, M. J., Piddock, L. J. V., & Webber, M. A. (2007). Commonly used farm disinfectants can select for mutant Salmonella enterica serovar Typhimurium with decreased susceptibility to biocides and antibiotics without compromising virulence. Journal of Antimicrobial Chemotherapy, 60(6), 1273–1280. https://doi.org/10.1093/jac/dkm359
Rensing, C., Moodley, A., Cavaco, L. M., & McDevitt, S. F. (2018). Resistance to metals used in agricultural production. Microbiology Spectrum, 6(2), 6-2. https://doi.org/10.1128/microbiolspec.ARBA-0025-2017
Rhouma, M., Romero-Barrios, P., Gaucher, M. L., & Bhachoo, S. (2021). Antimicrobial resistance associated with the use of antimicrobial processing aids during poultry processing operations: cause for concern? Critical Reviews in Food Science and Nutrition, 61(19), 3279-3296. https://doi.org/10.1080/10408398.2020.1798345
Roedel, A., Vincze, S., Projahn, M., Roesler, U., Robé, C., Hammerl, J. A., Noll, M., Dahouk, S. A., & Dieckmann, R. (2021). Genetic but no phenotypic associations between biocide tolerance and antibiotic resistance in Escherichia coli from German broiler fattening farms. Microorganisms, 9(3), 651. https://doi.org/10.3390/microorganisms9030651
Romero, J. L., Grande Burgos, M. J., Perez-Pulido, R., Galvez, A., & Lucas, R. (2017). Resistance to antibiotics, biocides, preservatives and metals in bacteria isolated from Seafoods: co-selection of strains resistant or tolerant to different classes of compounds. Fronters in Microbiology, 8, 1650. https://doi.org/10.3389/fmicb.2017.01650
Rossi, F., Rizzotti, L., Felis, G. E., & Torriani, S. (2014). Horizontal gene transfer among microorganisms in food: current knowledge and future perspectives. Food Microbiology, 42, 232-243. https://doi.org/10.1016/j.fm.2014.04.004
Scientific Committee on Emerging and Newly Identified Health Risks (SCENIHR) (2009). Assessment of the antibiotic resistance effects of biocides. (Last accessed: April 2023)
Scientific Committee on Emerging and Newly Identified Health Risks (SCENIHR) (2010). Research strategy to address the knowledge gaps on the antimicrobial resistance effects of biocides. (Last accessed: April 2023)
Seiler, C., & Berendonk, T. U. (2012). Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Frontiers in Microbiology, 3, 399. https://doi.org/10.3389/fmicb.2012.00399
Shelton, N. W., Jacob, M. E., Tokach, M. D., Nelssen, J. L., Goodband, R. D., Dritz, S. S., de Rouchey, J. M., Amachawadi, R. G.,
Shi, X., & Nagaraja, T. G. (2009). Effects of copper sulfate, zinc oxide, and neoterramycin on weanling pig growth and antibiotic resistance rate for fecal Escherichia coli. In Kansas State University Swine Day 2009. Report of Progress 1020; Goodband, B., Tokach, M., Dritz, S., de Rouchey, J., Eds.; Kansas State University: Manhattan, KS, USA, 2009; pp. 73–79. (Last accessed January 2023)
Silbergeld, E. K., & Nachman, K. (2008). The environmental and public health risks associated with arsenical use in animal feeds. Annals of the New York Academy of Sciences, 1140(1), 346-357. https://doi.org/10.1196/annals.1454.049
Singer, A. C., Shaw, H., Rhodes, V., & Hart, A. (2016). Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Frontiers in Microbiology, 7, 1728. https://doi.org/10.3389/fmicb.2016.01728
Slifierz, M. J., Friendship, R. M., & Weese, J. S. (2015b). Methicillin-resistant Staphylococcus aureus in commercial swine herds is associated with disinfectant and zinc usage. Applied and Environmental Microbiology, 81(8), 2690–2695. https://doi.org/10.1128/AEM.00036-15
Slifierz, M. J., Friendship, R., & Weese, J. S. (2015a). Zinc oxide therapy increases prevalence and persistence of methicillin-resistant Staphylococcus aureus in pigs: A randomized controlled trial. Zoonoses Public Health, 62, 301–308. https://doi.org/10.1111/zph.12150
Soumet, C., Fourreau, E., Legrandois, P., & Maris, P. (2012). Resistance to phenicol compounds following adaptation to quaternary ammonium compounds in Escherichia coli. Veterinary Microbiology, 158(1-2), 147-152. https://doi.org/10.1016/j.vetmic.2012.01.030
Stevanović, Z. D., Bošnjak-Neumüller, J., Pajić-Lijaković, I., Raj, J., & Vasiljević, M. (2018). Essential oils as feed additives—Future perspectives. Molecules, 23(7), 1717. https://doi.org/10.3390/molecules23071717
Tchounwou, P. B., Yedjou, C. G., Patlolla, A. K., & Sutton, D. J. (2012). Heavy metal toxicity and the environment. Molecular, Clinical and Environmental Toxicology: Volume 3: Environmental Toxicology, 133-164. https://doi.org/10.1007/978-3-7643-8340-4_6
Thomas, L., Maillard, J. Y., Lambert, R. J. W., & Russell, A. D. (2000). Development of resistance to chlorhexidine diacetate in Pseudomonas aeruginosa and the effect of a 'residual' concentration. Journal of Hospital Infection, 46(4), 297-303. https://doi.org/10.1053/jhin.2000.0851
Tongyi, Y., Yanpeng, L., Xingang, W., Fen, Y., Jun, L., & Yubin, T. (2020). Co‐selection for antibiotic resistance genes is induced in a soil amended with zinc. Soil Use and Management, 36(2), 328-337. https://doi.org/10.1111/sum.12545
Toyofuku, M., Nomura, N., & Eberl, L. (2019). Types and origins of bacterial membrane vesicles. Nature Reviews Microbiology, 17(1), 13-24. https://doi.org/10.1038/s41579-018-0112-2
Tu, Z., Shui, J., Liu, J., Tuo, H., Zhang, H., Lin, C., Feng, J., Feng, Y., Su, W. & Zhang, A. (2023). Exploring the abundance and influencing factors of antimicrobial resistance genes in manure plasmidome from swine farms. Journal of Environmental Sciences, 124, 462-471. https://doi.org/10.1016/j.jes.2021.11.030
Turchi, B., Bertelloni, F., Marzoli, F., Cerri, D., Tola, S., Azara, E., Longheu, C. M., Tassi, R., Schiavo, M., Cilia, G. & Fratini, F. (2020). Coagulase negative staphylococci from ovine milk: Genotypic and phenotypic characterization of susceptibility to antibiotics, disinfectants and biofilm production. Small Ruminant Research, 183, 106030. https://doi.org/10.1016/j.smallrumres.2019.106030
Uddin, M. J., Dawan, J., Jeon, G., Yu, T., He, X., & Ahn, J. (2020). The role of bacterial membrane vesicles in the dissemination of antibiotic resistance and as promising carriers for therapeutic agent delivery. Microorganisms, 8(5), 670. https://doi.org/10.3390/microorganisms8050670
Uruén, C., Chopo-Escuin, G., Tommassen, J., Mainar-Jaime, R. C., & Arenas, J. (2021). Biofilms as promoters of bacterial antibiotic resistance and tolerance. Antibiotics, 10(1), 3. https://doi.org/10.3390/antibiotics10010003
van Alen, S., Kaspar, U., Idelevich, E. A., Köck, R., & Becker, K. (2018). Increase of zinc resistance in German human derived livestock-associated MRSA between 2000 and 2014. Veterinary microbiology, 214, 7-12. https://doi.org/10.1016/j.vetmic.2017.11.032
Van Noten, N., Gorissen, L., & De Smet, S. (2016). Assistance in the Update of the Systematic Literature Review (SLR): “Influence of Copper on Antibiotic Resistance of Gut Microbiota on Pigs (including Piglets)”. EFSA supporting publication, 2016:EN-1005. 125 pp. https://doi.org/10.2903/sp.efsa.2016.EN-1005
Vats, P., Kaur, U. J., & Rishi, P. (2022). Heavy metal‐induced selection and proliferation of antibiotic resistance: A review. Journal of Applied Microbiology, 132(6), 4058-4076. https://doi.org/10.1111/jam.15492
Verraes, C., Van Boxstael, S., Van Meervenne, E., Van Coillie, E., Butaye, P., Catry, B., De Schaetzen, M. A., Van Huffel, X, Imberechts, H., Dierick, K., Daube, G., Saegerman, C., De Block, J., Dewulf, J., & Herman, L. (2013). Antimicrobial resistance in the food chain: a review. International Journal of Environmental Research and Public Health, 10(7), 2643-2669. https://doi.org/10.3390/ijerph10072643
VKM. (2016). Antimicrobial resistance due to the use of biocides and heavy metals: a literature review. Scientific Opinion on the Panel on Microbial Ecology of the Norwegian Scientific Committee for Food Safety, ISBN: 978-82-8259-253-6, Oslo, Norway. (Last accessed: January 2023)
Von Wintersdorff, C. J., Penders, J., Van Niekerk, J. M., Mills, N. D., Majumder, S., Van Alphen, L. B., Savelkoul, P. H. M., &
Wolffs, P. F. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Frontiers in Microbiology, 7, 173. https://doi.org/10.3389/fmicb.2016.00173
Wagner, T., Joshi, B., Janice, J., Askarian, F., Škalko-Basnet, N., Hagestad, O. C., Mekhkif, A., Wai, S. N., Hegstad, K., & Johannessen, M. (2018). Enterococcus faecium produces membrane vesicles containing virulence factors and antimicrobial resistance related proteins. Journal of Proteomics, 187, 28-38. https://doi.org/10.1016/j.jprot.2018.05.017
Wales, A. D., & Davies, R. H. (2015). Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics. 4(4):567–604. https://doi.org/10.3390/antibiotics4040567
Wang, H., Dong, Y., Yang, Y., Toor, G. S., & Zhang, X. (2013). Changes in heavy metal contents in animal feeds and manures in an intensive animal production region of China. Journal of Environmental Sciences, 25(12), 2435-2442. https://doi.org/10.1016/S1001-0742(13)60473-8
Wang, J., Wang, L., Zhu, L., Wang, J., & Xing, B. (2022a). Antibiotic resistance in agricultural soils: Source, fate, mechanism and attenuation strategy. Critical Reviews in Environmental Science and Technology, 52(6), 847-889. https://doi.org/10.1080/10643389.2020.1835438
Wang, M., Liu, P., Zhou, Q., Tao, W., Sun, Y., & Zeng, Z. (2018). Estimating the contribution of bacteriophage to the dissemination of antibiotic resistance genes in pig feces. Environmental Pollution, 238, 291-298. https://doi.org/10.1016/j.envpol.2018.03.024
Wang, Q., Awasthi, M. K., Zhang, Z., & Wong, J. W. (2019). Sustainable composting and its environmental implications. In Sustainable Resource Recovery and Zero Waste Approaches (pp. 115-132). Elsevier. https://doi.org/10.1016/B978-0-444-64200-4.00009-8
Wang, Q., Liu, L., Hou, Z., Wang, L., Ma, D., Yang, G., Guo, S., Luo, J., Qi, L., & Luo, Y. (2020). Heavy metal copper accelerates the conjugative transfer of antibiotic resistance genes in freshwater microcosms. Science of the Total Environment, 717, 137055. https://doi.org/10.1016/j.scitotenv.2020.137055
Wang, Z. F., Yun, H., Li, S., Ji, J., Khan, A., Fu, X. L., Zhang, P., & Li, X. K. (2022b). Factors influencing the transfer and abundance of antibiotic resistance genes in livestock environments in China. International Journal of Environmental Science and Technology, 1-12. https://doi.org/10.1007/s13762-022-04031-z
Watts, J. E., Schreier, H. J., Lanska, L., & Hale, M. S. (2017). The rising tide of antimicrobial resistance in aquaculture: sources, sinks and solutions. Marine Drugs, 15(6), 158. https://doi.org/10.3390/md15060158
Webber, M. A., & Piddock, L. J. V. (2003). The importance of efflux pumps in bacterial antibiotic resistance. Journal of Antimicrobial Chemotherapy, 51(1), 9-11. https://doi.org/10.4161/viru.23724
Weber, D. J., Rutala, W. A., & Sickbert-Bennett, E. E. (2019). Use of germicides in health care settings—is there a relationship between germicide use and antimicrobial resistance: A concise review. American Journal of Infection Control, 47, A106-A109. https://doi.org/10.1016/j.ajic.2019.03.023
Wesgate, R., Grasha, P., & Maillard, J. Y. (2016). Use of a predictive protocol to measure the antimicrobial resistance risks associated with biocidal product usage. American Journal of Infection Control, 44(4), 458-464. https://doi.org/10.1016/j.ajic.2015.11.009
Whitehead, R. N., Overton, T. W., Kemp, C. L., & Webber, M. A. (2011). Exposure of Salmonella enterica serovar Typhimurium to high level biocide challenge can select multidrug resistant mutants in a single step. PLoS One, 6(7), e22833. https://doi.org/10.1371/journal.pone.0022833
WHO (World Health Organisation) (2018a). Antimicrobial resistance. (Last accessed: April 2022)
WHO (World Health Organisation) (2018b). Critically Important Antimicrobials for Human Medicine. 6th Revision 2018. (Last accessed: July 2021)
Wieland, N., Boss, J., Lettmann, S., Fritz, B., Schwaiger, K., Bauer, J., & Hölzel, C. S. (2017). Susceptibility to disinfectants in antimicrobial‐resistant and‐susceptible isolates of Escherichia coli, Enterococcus faecalis and Enterococcus faecium from poultry–ESBL/AmpC‐phenotype of E. coli is not associated with resistance to a quaternary ammonium compound, DDAC. Journal of Applied Microbiology, 122(6), 1508-1517. https://doi:10.1111/jam.13440
Williams, O., Clark, I., Gomes, R. L., Perehinec, T., Hobman, J. L., Stekel, D. J., Hyde, R., Dodds, C. & Lester, E. (2019). Removal of copper from cattle footbath wastewater with layered double hydroxide adsorbents as a route to antimicrobial resistance mitigation on dairy farms. Science of the Total Environment, 655, 1139-1149. https://doi.org/10.1016/j.scitotenv.2018.11.330
Woegerbauer, M., Bellanger, X., & Merlin, C. (2020). Cell-Free DNA: An underestimated source of antibiotic resistance gene dissemination at the interface between human activities and downstream environments in the context of wastewater reuse. Frontiers in Microbiology, 11, 671. https://doi.org/10.3389/fmicb.2020.00671
Wohde, M., Berkner, S., Junker, T., Konradi, S., Schwarz, L., & Düring, R. A. (2016). Occurrence and transformation of veterinary pharmaceuticals and biocides in manure: a literature review. Environmental Sciences Europe, 28, 1-25. https://doi.org/10.1186/s12302-016-0091-8
Wu, N., Zhang, W., Xie, S., Zeng, M., Liu, H., Yang, J., Liu, X. & Yang, F. (2020). Increasing prevalence of antibiotic resistance genes in manured agricultural soils in northern China. Frontiers of Environmental Science & Engineering, 14, 1-12. https://doi.org/10.1007/s11783-019-1180-x
Xiao, X., Bai, L., Wang, S., Liu, L., Qu, X., Zhang, J., Xiao, Y., Tang, B., Li, Y., Yang, H., & Yang, H. (2022). Chlorine tolerance and cross-resistance to antibiotics in poultry-associated Salmonella isolates in China. Frontiers in Microbiology, 12, 833743. https://doi.org/10.3389/fmicb.2021.833743
Xue, H., Wu, Z., Li, L., Li, F., Wang, Y., & Zhao, X. (2015). Coexistence of heavy metal and antibiotic resistance within a novel composite staphylococcal cassette chromosome in a Staphylococcus haemolyticus isolate from bovine mastitis milk. Antimicrobial Agents and Chemotherapy, 59(9), 5788-5792. https://doi.org/10.1128/AAC.04831-14
Xue, J., Wu, J., Hu, Y., Sha, C., Yao, S., Li, P., Lin, K. & Cui, C. (2021). Occurrence of heavy metals, antibiotics, and antibiotic resistance genes in different kinds of land-applied manure in China. Environmental Science and Pollution Research, 28, 40011-40021. https://doi.org/10.1007/s11356-021-13307-9
Yang, H., Wei, S. H., Hobman, J. L., & Dodd, C. E. (2020b). Antibiotic and metal resistance in Escherichia coli isolated from pig slaughterhouses in the United Kingdom. Antibiotics, 9(11), 746. https://doi.org/10.3390/antibiotics9110746
Yang, S., Deng, W., Liu, S., Yu, X., Mustafa, G. R., Chen, S., He, L., Ao, X., Yang, Y., Zhou, K., Li, B., Han, X., Xu, X., & Zou, L. (2020a). Presence of heavy metal resistance genes in Escherichia coli and Salmonella isolates and analysis of resistance gene structure in E. coli E308. Journal of Global Antimicrobial Resistance, 21, 420-426. http://dx.doi.org/10.1016/j.jgar.2020.01.009
Yazdankhah, S., Rudi, K., & Bernhoft, A. (2014). Zinc and copper in animal feed–development of resistance and co-resistance to antimicrobial agents in bacteria of animal origin. Microbial Ecology in Health and Disease, 25(1), 25862. http://dx.doi.org/10.3402/mehd.v25.25862
Yazdankhah, S., Skjerve, E., & Wasteson, Y. (2018). Antimicrobial resistance due to the content of potentially toxic metals in soil and fertilizing products. Microbial Ecology in Health and Disease, 29(1), 1548248. https://doi.org/10.1080/16512235.2018.1548248
Yu, H. R., Li, L. Y., Shan, L. L., Gao, J., Ma, C. Y., & Li, X. (2021). Effect of supplemental dietary zinc on the growth, body composition and anti-oxidant enzymes of coho salmon (Oncorhynchus kisutch) alevins. Aquaculture Reports, 20, 100744. https://doi.org/10.1016/j.aqrep.2021.100744
Yu, Z., Gunn, L., Wall, P., & Fanning, S. (2017). Antimicrobial resistance and its association with tolerance to heavy metals in agriculture production. Food Microbiology, 64, 23–32. https://doi.org/10.1016/j.fm.2016.12.009
Yuan, W., Tian, T., Yang, Q., & Riaz, L. (2020). Transfer potentials of antibiotic resistance genes in Escherichia spp. strains from different sources. Chemosphere, 246, 125736. https://doi.org/10.1016/j.chemosphere.2019.125736
Zalewska, M., Błażejewska, A., Czapko, A., & Popowska, M. (2021). Antibiotics and antibiotic resistance genes in animal manure–consequences of its application in agriculture. Frontiers in Microbiology, 12, 610656. https://doi.org/10.3389/fmicb.2021.610656
Zhang, Y. J., Hu, H. W., Gou, M., Wang, J. T., Chen, D., & He, J. Z. (2017b). Temporal succession of soil antibiotic resistance genes following application of swine, cattle and poultry manures spiked with or without antibiotics. Environmental Pollution, 231, 1621-1632. https://doi.org/10.1016/j.envpol.2017.09.074
Zhang, Y., Gu, A. Z., Cen, T., Li, X., He, M., Li, D., & Chen, J. (2018). Sub-inhibitory concentrations of heavy metals facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes in water environment. Environmental Pollution, 237, 74-82. https://doi.org/10.1016/j.envpol.2018.01.032
Zhang, Y., Gu, A. Z., He, M., Li, D., & Chen, J. (2017a). Subinhibitory concentrations of disinfectants promote the horizontal transfer of multidrug resistance genes within and across genera. Environmental Science & Technology, 51(1), 570–580. https://doi.org/10.1021/acs.est.6b03132
Zhou, B., Wang, C., Zhao, Q., Wang, Y., Huo, M., Wang, J., & Wang, S. (2016). Prevalence and dissemination of antibiotic resistance genes and coselection of heavy metals in Chinese dairy farms. Journal of Hazardous Materials, 320, 10-17. http://dx.doi.org/10.1016/j.jhazmat.2016.08.007
Zhou, Q., Wang, M., Zhong, X., Liu, P., Xie, X., Wangxiao, J., & Sun, Y. (2019). Dissemination of resistance genes in duck/fish polyculture ponds in Guangdong Province: correlations between Cu and Zn and antibiotic resistance genes. Environmental Science and Pollution Research, 26, 8182-8193. https://doi.org/10.1007/s11356-018-04065-2
Zingali, T., Chapman, T. A., Webster, J., Roy Chowdhury, P., & Djordjevic, S. P. (2020). Genomic characterisation of a multiple drug resistant IncHI2 ST4 plasmid in Escherichia coli ST744 in Australia. Microorganisms, 8(6), 896. https://doi.org/10.3390/microorganisms8060896
Zou, L., Meng, J., McDermott, P. F., Wang, F., Yang, Q., Cao, G., Hoffmann, M., & Zhao, S. (2014). Presence of disinfectant resistance genes in Escherichia coli isolated from retail meats in the USA. Journal of Antimicrobial Chemotherapy, 69(10), 2644–2649. https://doi.org/10.1093/jac/dku197.
Revision log
Published: 21 August 2023
Last updated: 6 September 2023